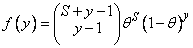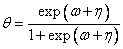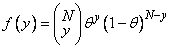.p>.CSCH5.DOC!CATEGORICAL_GLM;categorical_glm
Analyzes categorical data using logistic, Probit, Poisson, and other generalized linear models.
Synopsis
#include <imsls.h>
int imsls_f_categorical_glm (int n_observations, int n_class, int n_continuous, int model, float x[], ..., 0)
The type double function is imsls_d_categorical_glm.
Required Arguments
int
n_observations (Input)
Number of
observations.
int n_class
(Input)
Number
of classification variables.
int
n_continuous (Input)
Number of continuous
variables.
int model
(Input)
Argument model specifies
the model used to analyze the data. The six models are as follows:
|
Model |
Relationship* |
PDF of Response Variable |
|
0 |
Exponential |
Poisson |
|
1 |
Logistic |
Negative Binomial |
|
2 |
Logistic |
Logarithmic |
|
3 |
Logistic |
Binomial |
|
4 |
Probit |
Binomial |
|
5 |
Log-log |
Binomial |
Note that the lower bound of the response variable is 1 for model = 3 and is 0 for all other models. See the “Description” section for more information about these models.
float
x[] (Input)
Array of size
n_observations by
(n_class + n_continuous) + m
containing data for the independent variables, dependent variable, and optional
parameters.
The columns must be ordered such that the first n_class columns contain data for the class variables, the next n_continuous columns contain data for the continuous variables, and the next column contains the response variable. The final (and optional) m − 1 columns contain the optional parameters.
Return Value
An integer value indicating the number of estimated
coefficients
(n_coefficients)
in the model.
Synopsis with Optional Arguments
#include <imsls.h>
int
imsls_f_categorical_glm (int n_observations, int n_class,
int n_continuous, int
model,
float x[],
IMSLS_X_COL_DIM, int
x_col_dim,
IMSLS_X_COL_FREQUENCIES, int ifrq,
IMSLS_X_COL_FIXED_PARAMETER, int ifix,
IMSLS_X_COL_DIST_PARAMETER, int ipar,
IMSLS_X_COL_VARIABLES, int
iclass[],
int
icontinuous[], int iy,
IMSLS_EPS, float
eps,
IMSLS_MAX_ITERATIONS, int
max_iterations,
IMSLS_INTERCEPT,
IMSLS_NO_INTERCEPT,
IMSLS_EFFECTS, int
n_effects,
int
n_var_effects[],
int indices_effects,
IMSLS_INITIAL_EST_INTERNAL,
IMSLS_INITIAL_EST_INPUT, int
n_coef_input, float estimates[],
IMSLS_MAX_CLASS, int
max_class,
IMSLS_CLASS_INFO, int
**n_class_values, float **class_values,
IMSLS_CLASS_INFO_USER, int
n_class_values[], float class_values[],
IMSLS_COEF_STAT, float
**coef_statistics,
IMSLS_COEF_STAT_USER, float
coef_statistics[],
IMSLS_CRITERION, float
*criterion,
IMSLS_COV, float
**cov,
IMSLS_COV_USER, float
cov[],
IMSLS_MEANS, float
**means,
IMSLS_MEANS_USER, float
means[],
IMSLS_CASE_ANALYSIS, float
**case_analysis,
IMSLS_CASE_ANALYSIS_USER, float
case_analysis[],
IMSLS_LAST_STEP, float
**last_step,
IMSLS_LAST_STEP_USER, float
last_step[],
IMSLS_OBS_STATUS, int
**obs_status,
IMSLS_OBS_STATUS_USER, int
obs_status[],
IMSLS_ITERATIONS, int *n, float
**iterations,
IMSLS_ITERATIONS_USER, int *n, float
iterations[],
IMSLS_N_ROWS_MISSING, int
*n_rows_missing,
0)
Optional Arguments
IMSLS_X_COL_DIM, int x_col_dim
(Input)
Column dimension of input array x.
Default:
x_col_dim = n_class + n_continuous
+1
IMSLS_X_COL_FREQUENCIES, int ifrq
(Input)
Column number ifrg of x containing the
frequency of response for each observation.
IMSLS_X_COL_FIXED_PARAMETER, int ifix
(Input)
Column number ifix in x containing a fixed
parameter for each observation that is added to the linear response prior to
computing the model parameter. The ‘fixed’ parameter allows one to test
hypothesis about the parameters via the log-likelihoods.
IMSLS_X_COL_DIST_PARAMETER,
int
ipar (Input)
Column number ipar in x containing the value
of the known distribution parameter for each observation, where x[i][ipar] is the known
distribution parameter associated with the i-th observation. The meaning
of the distributional parameter depends upon model as follows:
|
model |
Parameter |
Meaning of x [i] [ipar] |
|
0 |
E |
ln (E) is a fixed intercept to be included in the linear predictor (i.e., the offset). |
|
1 |
S |
Number of successes required for the negative binomial distribution. |
|
2 |
- |
Not used for this model. |
|
3-5 |
N |
Number of trials required for the binomial distribution. |
Default: When model 2, each observation is assumed to have a parameter value of 1. When model = 2, this parameter is not referenced.
IMSLS_X_COL_VARAIBLES, int iclass[], int icontinuous[], int iy
(Input)
This keyword allows specification of the variables to be used
in the analysis and overrides the default ordering of variables described
for input argument x. Columns are
numbered 0 to x_col_dim_1. To avoid
errors, always specify the keyword IMSLS_X_COL_DIM when
using this keyword.
Argument iclass is an index vector of length n_class containing the column numbers of x that correspond to classification variables.
Argument icontinuous is an index vector of length n_continuous containing the column numbers of x that correspond to continuous variables.
Argument iy indicates the column of x which contains the independent variable.
IMSLS_EPS, float eps
(Input)
Argument eps is the convergence
criterion. Convergence is assumed when the maximum relative change in any
coefficient estimate is less than eps from one iteration
to the next or when the relative change in the log-likelihood, criterion, from
one iteration to the next is less than
eps / 100.0.
Default: eps = 0.001
IMSLS_MAX_ITERATIONS, int
max_iterations (Input)
Maximum number of iterations. Use
max_iterations = 0
to compute the Hessian, stored in cov, and the Newton
step, stored in last_step, at the
initial estimates (The initial estimates must be input. Use keyword IMSLS_INITIAL_EST_INPUT).
Default:
max_iterations = 30
IMSLS_INTERCEPT, or
IMSLS_NO_INTERCEPT,
By
default, or if IMSLS_INTERCEPT is
specified, the intercept is automatically included in the model. If IMSLS_NO_INTERCEPT is
specified, there is no intercept in the model (unless otherwise provided for by
the user).
IMSLS_EFFECTS, int n_effects, int n_var_effects[],
int indices_effects[]
(Input)
Variable n_effects is the
number of effects (sources of variation) in the model. Variable n_var_effects is an
array of length n_effects containing
the number of variables associated with each effect in the model. Argument indices_effects is an
index array of length n_var_effects [0] + n_var_effects [1] + … + n_var_effects [n_effects − 1]. The first n_var_effects [0]
elements give the column numbers of x for each variable in
the first effect. The next n_var_effects [1]
elements give the column numbers for each variable in the second effect.
The last n_var_effects [n_effects − 1] elements give the
column
numbers for each variable in the last effect.
IMSLS_INITIAL_EST_INTERNAL, or
IMSLS_INITIAL_EST_INPUT, int n_coef_input, float
estimates[] (Input)
By default, or if IMSLS_INIT_INTERNAL is
specified, then unweighted linear regression is used to obtain initial
estimates. If IMSLS_INITIAL_EST_INPUT
is specified, then the n_coef_input elements
of estimates
contain initial estimates of the parameters (which requires that the user know
the number of coefficients in the model prior to the call to imsls_f_categorical_glm
which can be obtained by calling imsls_f_regressors_for_glm.
IMSLS_MAX_CLASS, int max_class
(Input)
An upper bound on the sum of the number of distinct values taken on
by each classification variable.
Default: max_class = n_observations × n_class
IMSLS_CLASS_INFO, int
**n_class_values, float
**class_values (Output)
Argument n_class_values the
address of a pointer to the internally allocated array of length n_class containing the
number of values taken by each classification variable; the i-th
classification variable has n_class_values [i]
distinct values. Argument class_values is the
address of a pointer to the internally allocated array of length

containing the distinct values of the classification variables in ascending order. The first n_class_values [0] elements of class_values contain the values for the first classification variables, the next n_class_values [1] elements contain the values for the second classification variable, etc.
IMSLS_CLASS_INFO_USER, int
n_class_values[], float class_values[]
(Output)
Storage for arrays n_class_values and
class_values is
provided by the user. See IMSLS_CLASS_INFO.
IMSLS_COEF_STAT, float
**coef_statistics (Output)
Address of a pointer to an
internally allocated array of size n_coefficients × 4 containing the
parameter estimates and associated statistics, where n_coefficients can be
computed by calling imsls_regressors_for_glm.
|
Column |
Statistic |
|
0 |
Coefficient Estimate. |
|
1 |
Estimated standard deviation of the estimated coefficient. |
|
2 |
Asymptotic normal score for testing that the coefficient is zero. |
|
3 |
The p-value associated with the normal score in column 2. |
IMSLS_COEF_STAT_USER, float
coef_statistics[] (Output)
Storage for array coef_statistics is
provided by the user. See IMSLS_COEF_STAT.
IMSLS_CRITERION, float
*criterion (Output)
Optimized criterion. The criterion to
be maximized is a constant plus the log-likelihood.
IMSLS_COV, float **cov
(Output)
Address of a pointer to the internally allocated array of size n_coefficients × n_coefficients
containing the estimated asymptotic covariance matrix of the coefficients.
For max_iterations = 0,
this is the Hessian computed at the initial parameter estimates, where n_coefficients can be
computed by calling imsls_regressors_for_glm.
IMSLS_COV_USER, float cov[]
(Ouput)
Storage for array cov is provided by the
user. See IMSLS_COV
above.
IMSLS_MEANS, float **means
(Output)
Address of a pointer to the internally allocated array containing
the means of the design variables. The array is of length n_coefficients if
IMSLS_NO_INTERCEPT is
specified, and of length n_coefficients − 1 otherwise, where
n_coefficients
can be computed by calling imsls_regressors_for_glm.
IMSLS_MEANS_USER, float means[]
(Output)
Storage for array means is provided by the user. See IMSLS_MEANS.
IMSLS_CASE_ANALYSIS, float
**case_analysis (Output)
Address of a pointer to the
internally allocated array of size n_observations × 5 containing the case
analysis.
|
Column |
Statistic |
|
0 |
Predicted mean for the observation if model = 0. Otherwise, contains the probability of success on a single trial. |
|
1 |
The residual. |
|
2 |
The estimated standard error of the residual. |
|
3 |
The estimated influence of the observation. |
|
4 |
The standardized residual. |
Case statistics are computed for all observations except where missing values prevent their computation.
IMSLS_CASE_ANALYSIS_USER, float
case_analysis[] (Output)
Storage for array case_analysis is
provided by the user. See IMSLS_CASE_ANALYSIS.
IMSLS_LAST_STEP, float
**last_step (Output)
Address of a pointer to the
internally allocated array of length n_coefficients
containing the last parameter updates (excluding step halvings). For max_iterations = 0,
last_step
contains the inverse of the Hessian times the gradient vector, all computed
at the initial parameter estimates.
IMSLS_LAST_STEP_USER, float
last_step[] (Output)
Storage for array last_step is provided
by the user. See IMSLS_LAST_STEP.
IMSLS_OBS_STATUS, int
**obs_status (Output)
Address of a pointer to the
internally allocated array of length n_observations
indicating which observations are included in the extended likelihood.
|
obs_status [i] |
Status of observation |
|
0 |
Observation i is in the likelihood |
|
1 |
Observation i cannot be in the likelihood because it contains at least one missing value in x. |
|
2 |
Observation i is not in the likelihood. Its estimated parameter is infinite. |
IMSLS_OBS_STATUS_USER, int
obs_status[] (Output)
Storage for array obs_status is provided
by the user. See IMSLS_OBS_STATUS.
IMSLS_N_ROWS_MISSING, int
*n_rows_missing (Output)
Number of rows of data that
contain missing values in one or more of the following arrays or columns of
x; ipar, iy, ifrq, ifix, iclass, icontinuous, or indices_effects.
Remarks
1. Dummy variables are generated for the classification variables as follows: An ascending list of all distinct values of each classification variable is obtained and stored in class_values. Dummy variables are then generated for each but the last of these distinct values. Each dummy variable is zero unless the classification variable equals the list value corresponding to the dummy variable, in which case the dummy variable is one. See keyword IMSLS_LEAVE_OUT_LAST for optional argument IMSLS_DUMMY in routine imsls_f_regressors_for_glm (Chapter 2, “Regression”;).
2. The “product” of a classification variable with a covariate yields dummy variables equal to the product of the covariate with each of the dummy variables associated with the classification variable.
3. The “product” of two classification variables yields dummy variables in the usual manner. Each dummy variable associated with the first classification variable multiplies each dummy variable associated with the second classification variable. The resulting dummy variables are such that the index of the second classification variable varies fastest.
Description
Function imsls_f_categorical_glm uses iteratively reweighted least squares to compute (extended) maximum likelihood estimates in some generalized linear models involving categorized data. One of several models, including the probit, logistic, Poisson, logarithmic, and negative binomial models, may be fit.
Note that each row vector in the data matrix can represent a single observation; or, through the use of optional argument IMSLS_X_COL_FREQUENCIES, each row can represent several observations. Also note that classification variables and their products are easily incorporated into the models via the usual regression-type specifications.
The models available in imsls_f_categorical_glm
are:
|
Model |
PDF of the Response Variable |
Parameterization |
|
0 |
f (y) = (λy exp (−λ) ) / y! |
λ = N × exp ( + ) |
|
1 |
|
|
|
2 |
f (y) = (1 − θ)y / (yln θ) |
|
|
3 |
|
|
|
4 |
|
|
|
5 |
|
|
Here, denotes the cumulative normal distribution, N and S are known distribution parameters specified for each observation via the optional argument IMSLS_X_COL_DIST_PARAMETER, and is an optional fixed parameter of the linear response, i, specified for each observation. (If IMSLS_X_COL_FIXED_PARAMETER is not specified, then is taken to be 0.) Since the log-log model (model = 5) probabilities are not symmetric with respect to 0.5, quantitatively, as well as qualitatively, different models result when the definitions of “success” and “failure” are interchanged in this distribution. In this model and all other models involving θ, θ is taken to be the probability of a“success.”
Computational Details
The computations proceed as follows:
1. The input parameters are checked for consistency and validity.
2. Estimates of the means of the “independent” or design variables are computed. The frequency or the observation in all but binomial distribution models is taken from vector frequencies. In binomial distribution models, the frequency is taken as the product of n = parameter [i] and frequencies [i]. Means are computed as
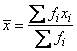
3. By default, and when IMSLS_INITIAL_EST_INTERNAL is specified, initial estimates of the coefficients are obtained (based upon the observation intervals) as multiple regression estimates relating transformed observation probabilities to the observation design vector. For example, in the binomial distribution models, θ may be estimated as
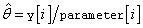
and, when model = 3, the linear relationship is given by
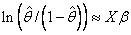
while if model = 4, Φ−1(θ) = Xβ. When computing initial estimates, standard modifications are made to prevent illegal operations such as division by zero. Regression estimates are obtained at this point, as well as later, by use of function imsls_f_regression (Chapter 2, “Regression”;).
4. Newton-Raphson iteration for the maximum likelihood estimates is implemented via iteratively re-weighted least squares. Let
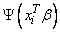
denote the log of the probability of the i-th observation for coefficients β. In the least-squares model, the weight of the i-th observation is taken as the absolute value of the second derivative of
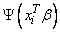
with respect to
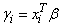
(times the frequency of the observation), and the dependent variable is taken as the first derivative Ψ with respect to γi, divided by the square root of the weight times the frequency. The Newton step is given by
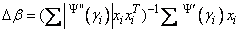
where all derivatives are evaluated at the current estimate of γ and βn+1 = β − Δβ. This step is computed as the estimated regression coefficients in the least-squares model. Step halving is used when necessary to ensure a decrease in the criterion.
5. Convergence is assumed when the maximum relative change in any coefficient update from one iteration to the next is less than eps or when the relative change in the log-likelihood from one iteration to the next is less than eps / 100. Convergence is also assumed after maxit iterations or when step halving leads to a step size of less than 0.0001 with no increase in the log-likelihood.
6. Residuals are computed according to methods discussed by Pregibon (1981). Let li (γi) denote the log-likelihood of the i-th observation evaluated at γi. Then, the standardized residual is computed as
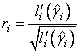
where
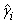
is the value of γi when evaluated at the optimal
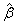
The denominator of this expression is used as the “standard error of the residual” while the numerator is “raw” residual. Following Cook and Weisberg (1982), the influence of the i-th observation is assumed to be
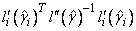
This quantity is a one-step approximation to the change in the estimates when the i-th observation is deleted. Here, the partial derivatives are with respect to β.
Programming Notes
1.
Indicator (dummy) variables are created for the classification variables using
function imsls_f_regressors_for_glm
(see Chapter 2, “Regression”;) using
keyword IMSLS_LEAVE_OUT_LAST
as the argument to the IMSLS_DUMMY optional
argument.
2. To enhance precision, “centering” of covariates is performed if the model has an intercept and n_observations − n_rows_missing > 1. In doing so, the sample means of the design variables are subracted from each observation prior to its inclusion in the model. On convergence, the intercept, its variance, and its covariance with the remaining estimates are transformed to the uncentered estimate values.
3. Two methods for specifying a binomial distribution model are possible. In the first method, frequencies contains the frequency of the observation while y is 0 or 1 depending upon whether the observation is a success or failure. In this case, N = parameter [i] is always 1. The model is treated as repeated Bernoulli trials, and interval observations are not possible. A second method for specifying binomial models is to use y to represent the number of successes in parameter [i] trials. In this case, frequencies will usually be 1.
Examples
Example 1
The first example is from Prentice (1976) and involves the mortality of beetles after five hours exposure to eight different concentrations of carbon disulphide. The table below lists the number of beetles exposed (N) to each concentration level of carbon disulphide (x, given as log dosage) and the number of deaths which result (y). The data is given as follows:
|
Log Dosage |
Number of Beetles Exposed |
Number of Deaths |
|
1.690 |
59 |
6 |
|
1.724 |
60 |
13 |
|
1.755 |
62 |
18 |
|
1.784 |
56 |
28 |
|
1.811 |
63 |
52 |
|
1.836 |
59 |
53 |
|
1.861 |
62 |
61 |
|
1.883 |
60 |
60 |
The number of deaths at each concentration level are fitted as a binomial response using logit (model = 3), probit (model = 4), and log-log (model = 5) models. Note that the log-log model yields a smaller absolute log likelihood (14.81) than the logit model (18.78) or the probit model (18.23). This is to be expected since the response curve of the log-log model has an asymmetric appearance, but both the logit and probit models are symmetric about θ = 0.5.
#include <imsls.h>
#include <stdio.h>
main ()
{
static float x[8][3] = { 1.69, 6, 59,
1.724, 13, 60,
1.755, 18, 62,
1.784, 28, 56,
1.811, 52, 63,
1.836, 53, 59,
1.861, 61, 62,
1.883, 60, 60};
float *coef_statistics, criterion;
int n_obs=8, n_class=0, n_continuous=1;
int n_coef, model=3, ipar=2;
char *fmt = "%12.4f";
static char *clabels[] = {"", "coefficients", "s.e", "z", "p"};
n_coef = imsls_f_categorical_glm (n_obs, n_class, n_continuous,
model, &x[0][0],
IMSLS_X_COL_DIST_PARAMETER, ipar,
IMSLS_COEF_STAT, &coef_statistics,
IMSLS_CRITERION, &criterion, 0);
imsls_f_write_matrix ("Coefficient statistics for model 3", n_coef, 4, coef_statistics,
IMSLS_WRITE_FORMAT, fmt, IMSLS_NO_ROW_LABELS, IMSLS_COL_LABELS, clabels,0);
printf ("\nLog likelihood %f \n", criterion);
model=4;
n_coef = imsls_f_categorical_glm (n_obs, n_class, n_continuous,
model, &x[0][0],
IMSLS_X_COL_DIST_PARAMETER, ipar,
IMSLS_COEF_STAT, &coef_statistics,
IMSLS_CRITERION, &criterion, 0);
imsls_f_write_matrix ("Coefficient statistics for model 4", n_coef, 4, coef_statistics,
IMSLS_WRITE_FORMAT, fmt, IMSLS_NO_ROW_LABELS, IMSLS_COL_LABELS, clabels,0);
printf ("\nLog likelihood %f \n", criterion);
model=5;
n_coef = imsls_f_categorical_glm (n_obs, n_class, n_continuous,
model, &x[0][0],
IMSLS_X_COL_DIST_PARAMETER, ipar,
IMSLS_COEF_STAT, &coef_statistics,
IMSLS_CRITERION, &criterion, 0);
imsls_f_write_matrix ("Coefficient statistics for model 5", n_coef, 4, coef_statistics,
IMSLS_WRITE_FORMAT, fmt, IMSLS_NO_ROW_LABELS, IMSLS_COL_LABELS, clabels,0);
printf ("\nLog likelihood %f \n", criterion);
}
Output
Coefficient statistics for model 3
coefficients s.e z p
-60.7568 5.1876 -11.7118 0.0000
34.2985 2.9164 11.7607 0.0000
Log likelihood -18.778181
Coefficient statistics for model 4
coefficients s.e z p
-34.9441 2.6412 -13.2305 0.0000
19.7367 1.4852 13.2888 0.0000
Log likelihood -18.232355
Coefficient statistics for model 5
coefficients s.e z p
-39.6133 3.2489 -12.1930 0.0000
22.0685 1.8047 12.2284 0.0000
Log likelihood -14.807850
Example 2
Consider the use of a loglinear model to analyze survival-time data. Laird and Oliver (1981) investigate patient survival post heart valve replacement surgery. Surveilance after surgery of the 109 patients included in the study ranged from 3 to 97 months. All patients were classified by heart valve type (aortic or mitral) and by age (less than 55 years or at least 55 years). The data could be considered as a three-way contingency table where patients are classified by valve type, age, and survival (yes or no). However, it would be inappropriate to analyze this data using the standard methodology associated with contingency tables; since, this methodology ignores survival time.
Consider a variable, say exposure time (Eij), that is defined as
the sum of the length of times patients of each cross-classification are at
risk. The length of time for a patient that dies is the number of months from
surgery until death and for a survivor, the length of time is the number of
months from surgery until the study ends or the patient withdraws from the
study. Now we can model the effect of
A = age and
V = valve type on the expected number of deaths conditional on
exposure time. Thus, for the data (shown in the table below), assume the number
of deaths are independent Poisson random variables with means mij and fit the following
model,
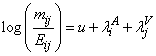
where u is the overall mean,
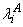
is the effect of age, and
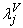
is the effect of the valve type.
|
|
|
Heart Valve Type | |
|
Age |
|
Aortic (0) |
Mitral (1) |
|
< 55 years (Age = 0) |
Deaths |
4 |
1 |
|
|
Exposure |
1259 |
2082 |
|
≥ 55 years (Age = 1) |
Deaths |
7 |
9 |
|
|
Exposure |
1417 |
1647 |
From the coefficient statistics table of the output, note that the risk is estimated to be e1.22 = 3.39 times higher for older patients in the study. This increase in risk is significant (p = 0.02). However, the decrease in risk for the mitral valve patients is estimated to be e−0.33 = 0.72 times that of the aortic valve patients and this risk is not significant (p = 0.45).
#include <imsls.h>
main ()
{
int nobs = 4;
int n_class = 2;
int n_cont = 0;
int model = 0;
float x[16] = {
4, 1259, 0, 0,
1, 2082, 0, 1,
7, 1417, 1, 0,
9, 1647, 1, 1
};
int iclass[2] = {2, 3};
int icont[1] = {-1};
int n_coef;
float *coef;
char *clabels[5] = {"", "coefficient", "std error", "z-statistic", "p- value"};
char *fmt = "%10.6W";
n_coef = imsls_f_categorical_glm(nobs, n_class, n_cont, model, x,
IMSLS_COEF_STAT, &coef,
IMSLS_X_COL_VARIABLES, iclass, icont, 0,
IMSLS_X_COL_DIST_PARAMETER, 1,
0);
imsls_f_write_matrix("Coefficient Statistics", n_coef, 4, coef,
IMSLS_COL_LABELS, clabels, IMSLS_ROW_NUMBER_ZERO,
IMSLS_WRITE_FORMAT, fmt, 0);
}
Output
Coefficient Statistics
coefficient std error z-statistic p-value
0 -5.4210 0.3456 -15.6837 0.0000
1 -1.2209 0.5138 -2.3763 0.0177
2 0.3299 0.4382 0.7528 0.4517
Warning Errors
IMSLS_TOO_MANY_HALVINGS Too many step halvings. Convergence is assumed.
IMSLS_TOO_MANY_ITERATIONS Too many iterations. Convergence is assumed.
Fatal Errors
IMSLS_TOO_FEW_COEF IMSLS_INITIAL_EST_INPUT is specified and “n_coef_input” = #. The model specified requires # coefficients.
IMSLS_MAX_CLASS_TOO_SMALL
The number of distinct values of the classification variables exceeds
“max_class”
= #.
IMSLS_INVALID_DATA_8 “n_class_values[#]” = #. The number of distinct values for each classification variable must be greater than one.
IMSLS_NMAX_EXCEEDED The number of observations to be deleted has exceeded “lp_max” = #. Rerun with a different model or increase the workspace.
*Relationship between the parameter, θ or λ, and a linear model of the explanatory variables, X β.
|
Visual Numerics, Inc. PHONE: 713.784.3131 FAX:713.781.9260 |