Computes cluster membership for a hierarchical cluster tree.
Synopsis
int *imsls_cluster_number (int npt, int *iclson, int *icrson, int k, …, 0)
Required Arguments
int npt
(Input)
Number of data points to be clustered.
int *iclson (Input)
Vector
of length npt - 1 containing the
left son cluster numbers.
Cluster npt + i is formed by merging
clusters iclson[i-1] and icrson[i-1].
int *icrson (Input)
Vector
of length npt - 1 containing the
left son cluster numbers.
Cluster npt + i is formed by merging
clusters iclson[i-1] and icrson[i-1].
int k
(Input)
Desired number of clusters.
Return Value
Vector of length npt containing the cluster membership of each observation.
Synopsis with Optional Arugments
int *imsls_cluster_number (int npt, int *iclson, int *icrson, int k,
IMSLS_OBS_PER_CLUSTERS, int
**nclus,
IMSLS_OBS_PER_CLUSTERS_USER, int nclus[],
IMSLS_RETURN_USER, int
iclus[],
0)
Optional Arguments
IMSLS_OBS_PER_CLUSTERS,
int **nclus
(Output)
Address of a pointer to an internally allocated array of
length k containing the
number of observations in each cluster.
IMSLS_OBS_PER_CLUSTERS_USER, int nclus[] (Output)
Storage
for array nclus is provided by
the user. See IMSLS_OBS_PER_CLUSTERS.
IMSLS_RETURN_USER, float
iclus[] (Output)
User allocated array of length npt containing the
cluster membership of each observation.
Description
Given a fixed number of clusters (K) and the cluster tree (vectors icrson and iclson) produced by the hierarchical clustering algorithm (see function imsls_f_cluster_hierarchical, function imsls_cluster_number determines the cluster membership of each observation. The function imsls_cluster_number first determines the root nodes for the K distinct subtrees forming the K clusters and then traverses each subtree to determine the cluster membership of each observation. The function imsls_cluster_number also returns the number of observations found in each cluster.
Example 1
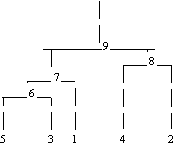
In the following example, cluster membership for K = 2 clusters is found for the displayed cluster tree. The output vector iclus contains the cluster numbers for each observation.
iclus = imsls_cluster_number(npt, iclson, icrson, k, 0);
imsls_i_write_matrix("iclus", 1, 5, iclus, 0);
Output
Example 2
This example illustrates the typical usage of imsls_cluster_number. The Fisher iris data (see function imsls_f_data_sets, see Chapter 15, “Utilities”.p<.STCH19.DOC!GDATA;;) is clustered. First the distance between the irises are computed using function imsls_f_dissimilarities. The resulting distance matrix is then clustered using function imsls_f_cluster_hierarchical. The cluster membership for 5 clusters is then obtained via function imsls_cluster_number using the output from imsls_f_cluster_hierarchical. The need for 5 clusters can be obtained either by theoretical means or by examining a cluster tree. The cluster membership for each of the iris observations is printed.
#define MAX(A,B) ((A)>(B)?(A): (B))
int ncol = 5, nrow = 150, nvar = 4, npt = 150, k = 5;
int i, j, *iclson, *icrson, *iclus, *nclus;
float *clevel, dist[150][150], *x, f_rand;
int *p_iclus = NULL, *p_nclus = NULL;
imsls_f_dissimilarities(nrow, ncol, x,
imsls_f_random_uniform (1, IMSLS_RETURN_USER, &f_rand, 0);
dist[i][j] = MAX (0.0, dist[i][j] + .001 * f_rand);
imsls_f_cluster_hierarchical (npt, (float*)dist,
IMSLS_CLUSTERS, &clevel, &iclson, &icrson,
iclus = imsls_cluster_number (npt, iclson, icrson, k,
IMSLS_OBS_PER_CLUSTER, &nclus,
imsls_i_write_matrix ("iclus", 25, 5, iclus, 0);
imsls_i_write_matrix ("nclus", 1, 5, nclus, 0); }
Output
|
Visual Numerics, Inc. PHONE: 713.784.3131 FAX:713.781.9260 |