Computes partial covariances or partial correlations from the covariance or correlation matrix.
Synopsis
#include <imsls.h>
float *imsls_f_partial_covariances (int n_independent, int n_dependent, float x, ..., 0)
The type double function is imsls_d_partial_covariances.
Required Argument
int
n_independent (Input)
Number of “independent” variables to
be used in the partial covariances/correlations. The partial
covariances/correlations are the covariances/correlations between the dependent
variables after removing the linear effect of the independent variables.
int
n_dependent (Input)
Number of variables for which partial
covariances/correlations are desired (the number of “dependent” variables).
float x
(Input)
The n × n covariance or
correlation matrix, where n = n_independent + n_dependent. The
rows/columns must be ordered such that the first n_independent
rows/columns contain the independent variables, and the last n_dependent
row/columns contain the dependent variables. Matrix x must always be
square symmetric.
Return Value
Matrix of size n_dependent by n_dependent containing the partial covariances (the default) or partial correlations (use keyword IMSLS_PARTIAL_CORR).
Synopsis with Optional Arguments
#include <imsls.h>
float
*imsls_f_partial_covariances (int n_independent, int n_dependent, float x[],
IMSLS_X_COL_DIM, int
x_col_dim,
IMSLS_X_INDICES, int
indices[],
IMSLS_PARTIAL_COV, or
IMSLS_PARTIAL_CORR,
IMSLS_TEST, int df, int
*df_out,
float
**p_values,
IMSLS_TEST_USER, int df,
int *df_out, float
p_values[],
IMSLS_RETURN_USER, float
c[],
0)
Optional Arguments
IMSLS_X_COL_DIM, int x_col_dim
(Input)
Row/Column dimension of x.
Default:
x_col_dim = n_independent + n_dependent.
IMSLS_X_INDICES, int indices[]
(Input)
An array of length x_col_dim containing
values indicating the status of the variable as in the following table:
|
indices[i] |
Variable is... |
|
−1 |
not used in analysis |
|
0 |
dependent variable |
|
1 |
independent variable |
By default, the first n_independent elements of indices are equal to 1, and the last n_dependent elements are equal to 0.
IMSLS_PARTIAL_COV, or
IMSLS_PARTIAL_CORR,
By
default, and if IMSLS_PARTIAL_COV is
specified, partial covariances are calculated. Partial correlations are
calculated if IMSLS_PARTIAL_CORR is
specified.
IMSLS_TEST, int df, int *df_out, float
**p_values
(Input,
Output, Output)
Argument df is an input integer
indicating the number of degrees of freedom associated with input matrix
x. If the number
of degrees of freedom in x varies from element
to element, then a conservative choice for df is the minimum
degrees of freedom for all elements in x.
Argument df_out contains the number of degrees of freedom in the test that the partial covariances/correlations are zero. This value will usually be df − n_independent, but will be greater than this value if the independent variables are computationally linearly related.
Argument p_values is the address of a pointer to an internally allocated array of size n_dependent by n_dependent containing the p-values for testing the null hypothesis that the associated partial covariance/correlation is zero. It is assumed that the observations from which x was computed flows a multivariate normal distribution and that each element in x has df degrees of freedom.
IMSLS_TEST_USER, int df, int *df_out, float
p_values[]
(Input,
Output, Output)
Storage for array p_values is provided
by the user. See IMSLS_TEST
above.
IMSLS_RETURN_USER, float c[]
(Output)
If specified, c returns the partial
covariances/correlations. Storage for array c is provided by
the user.
Description
Function imsls_f_partial_covariances computed partial covariances or partial correlations from an input covariance or correlation matrix. If the “independent” variables (the linear “effect” of the independent variables is removed in computing the partial covariances/correlations) are linearly related to one another, imsls_f_partial_covariances detects the linearity and eliminates one or more of the independent variables from the list of independent variables. The number of variables eliminated, if any, can be determined from argument df_out.
Given a covariance or correlation matrix Σ partitioned as
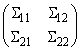
function imsls_f_partial_covariances computed the partial covariances (of the standardized variables if Σ is a correlation matrix) as
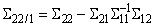
If partial correlations are desired, these are computed as
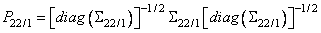
where diag denotes the matrix containing the diagonal of its argument along its diagonal with zeros off the diagonal. If Σ 11 is singular, then as many variables as required are deleted from Σ 11 (and Σ 12) in order to eliminate the linear dependencies. The computations then proceed as above.
The p-value for a partial covariance tests the null hypothesis H0: σ ij|1 = 0, where σij|1 is the (i, j) element in matrix Σ22|1. The p-value for a partial correlation tests the null hypothesis H0: ρij|1 = 0, where ρij|1 is the (i, j) element in matrix P22|1. The p-values are returned in p_values. If the degrees of freedom for x, df, is not known, the resulting p-values may be useful for comparison, but they should not by used as an approximation to the actual probabilities.
Examples
Example 1
The following example computes partial covariances, scaled from a nine-variable correlation matrix originally given by Emmett (1949). The first three rows and columns contain the independent variables and the final six rows and columns contain the dependent variables.
#include <imsls.h>
#include <math.h>
main()
{
float *pcov;
float x[9][9] = {
6.300, 3.050, 1.933, 3.365, 1.317, 2.293, 2.586, 1.242, 4.363,
3.050, 5.400, 2.170, 3.346, 1.473, 2.303, 2.274, 0.750, 4.077,
1.933, 2.170, 3.800, 1.970, 0.798, 1.062, 1.576, 0.487, 2.673,
3.365, 3.346, 1.970, 8.100, 2.983, 4.828, 2.255, 0.925, 3.910,
1.317, 1.473, 0.798, 2.983, 2.300, 2.209, 1.039, 0.258, 1.687,
2.293, 2.303, 1.062, 4.828, 2.209, 4.600, 1.427, 0.768, 2.754,
2.586, 2.274, 1.576, 2.255, 1.039, 1.427, 3.200, 0.785, 3.309,
1.242, 0.750, 0.487, 0.925, 0.258, 0.768, 0.785, 1.300, 1.458,
4.363, 4.077, 2.673, 3.910, 1.687, 2.754, 3.309, 1.458, 7.400};
pcov = imsls_f_partial_covariances(3, 6, x, 0);
imsls_f_write_matrix("Partial Covariances", 6, 6, pcov, 0);
free(pcov);
return;
}
Output
Partial Covariances
1 2 3 4 5 6
1 0.000 0.000 0.000 0.000 0.000 0.000
2 0.000 0.000 0.000 0.000 0.000 0.000
3 0.000 0.000 0.000 0.000 0.000 0.000
4 0.000 0.000 0.000 5.495 1.895 3.084
5 0.000 0.000 0.000 1.895 1.841 1.476
6 0.000 0.000 0.000 3.084 1.476 3.403
Example 2
The following example computes partial correlations from a
9 variable correlation matrix originally given by Emmett (1949). The partial
correlations between the remaining variables, after adjusting for variables 1, 3
and 9, are computed. Note in the output that the row and column labels are
numbers, not variable numbers. The corresponding variable numbers would be 2, 4,
5, 6, 7
and 8, respectively.
#include <imsls.h>
main()
{
float *pcorr, *pval;
int df;
float x[9][9] = {
1.0, 0.523, 0.395, 0.471, 0.346, 0.426, 0.576, 0.434, 0.639,
0.523, 1.0, 0.479, 0.506, 0.418, 0.462, 0.547, 0.283, 0.645,
0.395, 0.479, 1.0, .355, 0.27, 0.254, 0.452, 0.219, 0.504,
0.471, 0.506, 0.355, 1.0, 0.691, 0.791, 0.443, 0.285, 0.505,
0.346, 0.418, 0.27, 0.691, 1.0, 0.679, 0.383, 0.149, 0.409,
0.426, 0.462, 0.254, 0.791, 0.679, 1.0, 0.372, 0.314, 0.472,
0.576, 0.547, 0.452, 0.443, 0.383, 0.372, 1.0, 0.385, 0.68,
0.434, 0.283, 0.219, 0.285, 0.149, 0.314, 0.385, 1.0, 0.47,
0.639, 0.645, 0.504, 0.505, 0.409, 0.472, 0.68, 0.47, 1.0};
int indices[9] = {1, 0, 1, 0, 0, 0, 0, 0, 1};
pcorr = imsls_f_partial_covariances(3, 6, &x[0][0],
IMSLS_PARTIAL_CORR,
IMSLS_X_INDICES, indices,
IMSLS_TEST, 30, &df, &pval,
0);
printf ("The degrees of freedom are
%d\n\n", df);
imsls_f_write_matrix("Partial Correlations", 6, 6, pcorr, 0);
imsls_f_write_matrix("P-Values", 6, 6, pval, 0);
free(pcorr);
free(pval);
return;
}
Output
The degrees of freedom are 27
Partial Correlations
1 2 3 4 5 6
1 1.000 0.224 0.194 0.211 0.125 -0.061
2 0.224 1.000 0.605 0.720 0.092 0.025
3 0.194 0.605 1.000 0.598 0.123 -0.077
4 0.211 0.720 0.598 1.000 0.035 0.086
5 0.125 0.092 0.123 0.035 1.000 0.062
6 -0.061 0.025 -0.077 0.086 0.062 1.000
P-Values
1 2 3 4 5 6
1 0.0000 0.2525 0.3232 0.2801 0.5249 0.7576
2 0.2525 0.0000 0.0006 0.0000 0.6417 0.9000
3 0.3232 0.0006 0.0000 0.0007 0.5328 0.6982
4 0.2801 0.0000 0.0007 0.0000 0.8602 0.6650
5 0.5249 0.6417 0.5328 0.8602 0.0000 0.7532
6 0.7576 0.9000 0.6982 0.6650 0.7532 0.0000
Warning Errors
IMSLS_NO_HYP_TESTS The input matrix “x” has # degrees of freedom, and the rank of the dependent variables is #. There are not enough degrees of freedom for hypothesis testing. The elements of “p_values” are set to NaN (not a number).
Fatal Errors
IMSLS_INVALID_MATRIX_1 The input matrix “x” is incorrectly specified. A computed correlation is greater than 1 for variables # and #.
IMSLS_INVALID_PARTIAL A computed partial correlation for variables # and # is greater than 1. The input matrix “x” is not positive semi-definite.
|
Visual Numerics, Inc. PHONE: 713.784.3131 FAX:713.781.9260 |