Estimates missing observations in designed experiments using Yate’s method.
Synopsis
#include <imsls.h>
int imsls_f_yates(int n, int n_independent, float x[],…, 0)
The type double function is imsls_d_yates.
Required Arguments
int n (Input)
Number of
observations.
int n_independent
(Input)
Number of independent variables.
float x[]
(Input/Output)
A n by (n_independent+1)
2-dimensional array containing the experimental observations and missing
values. The first n_independent columns
contain values for the independent variables and the last column contains the
corresponding observations for the dependent variable or response. The
columns assigned to the independent variables should not contain any missing
values. Missing values are included in this array by placing a NaN (not a number) in
the last column of x. The NaN value can be set
using either the function imsls_f_machine(6) or
imsls_d_machine(6),
depending upon whether single or double precision is being used,
respectively. Upon successful completion, missing values are replaced with
estimates calculated using Yates’ method.
Return Value
The number of missing values replaced with estimates using the Yates procedure. A negative return value indicates that the routine was unable to successfully estimate all missing values. Typically this occurs when all of the observations for a particular treatment combination are missing. In this case, Yate’s missing value method does not produce a unique set of missing value estimates.
Synopsis with Optional Arugments
#include <imsls.h>
int
imsls_f_yates (int
n, int n_independent, float
x[],
IMSLS_DESIGN, int
design,
IMSLS_INITIAL_ESTIMATES, int
n_missing,
float
initial_estimates[],
IMSLS_GET_SS, float
get_ss (int n, int
n_independent,
int n_levels[], float
dataMatrix[]),
IMSLS_GRAD_TOL, float
grad_tol,
IMSLS_STEP_TOL, float
step_tol,
IMSLS_MAX_ITN, int
**itmax,
IMSLS_MISSING_INDEX, int
**missing_index[],
IMSLS_MISSING_INDEX_USER, int
missing_index[],
IMSLS_ERROR_SS, float
*error_ss,
0)
Optional Arguments
IMSLS_RETURN_USER,
int n_missing (Output)
The number of missing values replaced with
Yate’s estimates. A negative return value indicates that the routine was
unable to successfully estimate all missing values.
IMSLS_DESIGN,
int design
(Input)
An integer indicating whether a custom or standard design is being
used. The association of values for this variable and standard designs is
described in the following table:
|
Design |
Description |
|
0 |
CRD – Completely Randomized Design. The input matrix, x, is assumed to have only two columns. The first is used to contain integers identifying the treatments. The second column should contain corresponding observations for the dependent variable. In this case, n_independent=1. Default value when n_independent=1. |
|
1 |
RCBD – Randomized Complete Block Design. The input matrix is assumed to have only three columns. The first is used to contain the treatment identifiers and the second the block identifiers. The last column contains the corresponding observations for the dependent variable. In this case, n_independent=2. This is the default value when n_independent=2. |
|
2 |
Another design. In this case, the function get_ss is a required input. The design matrix is passed to that routine. Initial values for missing observations are set to the grand mean of the data, unless initial values are specified using IMSLS_INITIAL_ESTIMATES. |
Default: design=0 or design=1, depending upon whether n_independent=1 or 2 respectively. If n_independent>2, then design must be set to 2, and get_ss must be provided as input to imsls_f_yates.
IMSLS_INITIAL_ESTIMATES,
int n_missing,
float
initial_estimates[] (Input)
Initial estimates for
the missing values. Argument n_missing is the
number of missing values. Argument initial_estimates is
an array of length n_missing containing
the initial estimates.
Default: For design=0 and design=1, the initial
estimates are calculated using the Yates formula for those designs. For design=2, the mean of
the non-missing observations is used as the initial estimate for all missing
values.
IMSLS_MAX_ITN,
int itmax
(Input)
Maximum number of iterations in the optimization routine for
finding the missing value estimates that minimize the error sum of squares in
the analysis of variance.
Default: itmax = 500.
IMSLS_GET_SS,
float get_ss(int
n,
int
n_independent, int n_levels[],
float
dataMatrix[]) (Input/Output)
A user-supplied function
that returns the error sum of squares calculated using the n by (n_independent+1)
matrix dataMatrix.
imsls_f_yates
calculates the error sum of squares assuming that dataMatrix contains no
missing observations. In general, dataMatrix
should be equal to the input matrix x with missing values
replaced by estimates. imsls_f_yates is
required input when design=2. The array
n_levels should
be of length n_independent and
contain the number of levels associated with each of the first n_independent columns
in the dataMatrix and x arrays.
IMSLS_GRAD_TOL,
float grad_tol
(Input)
Scaled gradient tolerance used to determine whether the difference
between the error sum of squares is small enough to stop the search for missing
value estimates.
Default: grad_tol = 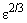 , where
, where 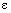 is the machine precision.
is the machine precision.
IMSLS_STEP_TOL,
float step_tol
(Input)
Scaled step tolerance used to determine whether the difference
between missing value estimates is small enough to stop the search for missing
value estimates.
Default: step_tol = 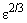 , where
, where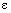 is the machine precision.
is the machine precision.
IMSLS_MISSING_INDEX,
int *missing_index
(Output)
An array of length n_missing containing
the indices for the missing values in x. The number of
missing values, n_missing, is the
return value of imsls_f_yates.
IMSLS_MISSING_INDEX_USER,
int missing_index[]
(Output)
Storage for the array missing_index,
provided by the user.
IMSLS_ERROR_SS,
float *errr_ss
(Output)
The value of the error sum of squares calculated using the
missing value estimates. If design=2 then this is
equal to the value returned from get_ss using the Yates
missing value estimates.
Description
Several functions for analysis of variance require balanced experimental data, i.e. data containing no missing values within a block and an equal number of replicates for each treatment. If the number of missing observations in an experiment is smaller than the Yates method as described in Yates (1933) and Steel and Torrie (1960), can be used to estimate the missing values. Once the missing values are replaced with these estimates, the data can be passed to an analysis of variance that requires balanced data.
The basic principle behind the Yates method for estimating missing observations is to replace the missing values with values that minimize the error sum of squares in the analysis of variance. Since the error sum of squares depends upon the underlying model for the analysis of variance, the Yates formulas for estimating missing values vary from anova to anova.
Consider, for example, the model underlying experiments
conducted using a completely randomized design. If 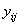 is the Ith observation for
the ith treatment then the error sum of squares for a CRD is calculated using
the following formula:
is the Ith observation for
the ith treatment then the error sum of squares for a CRD is calculated using
the following formula:
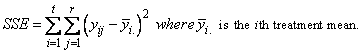
If an observation 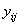 is missing then SSE is minimized by replacing that missing
observation with the estimate
is missing then SSE is minimized by replacing that missing
observation with the estimate
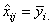 .
.
For a randomized complete block design (RCBD), the calculation for estimating a single missing observation can be derived from the RCBD error sum of squares:
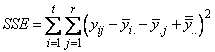
If only a single observation, 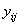 , is missing from the jth
block and ith treatment, the estimate for this missing observation can be
derived by solving the equation:
, is missing from the jth
block and ith treatment, the estimate for this missing observation can be
derived by solving the equation:
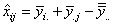 .
.
The solution is referred to as the Yates formula for a RCBD:
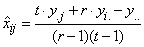 , where
, where
r=n_blocks,
t=n_treatments,
yi=total of all
non-missing observations from the ith treatment, 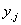 =total of all non-missing
observations from the jth block, and y=total of all non-missing
observations.
=total of all non-missing
observations from the jth block, and y=total of all non-missing
observations.
If more than one observation is missing, imsls_f_yates
minimization procedure is used to estimate missing values. For a CRD, all
missing observations are set equal to their corresponding treatment means
calculated using the non-missing observations. That is, 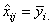 .
.
For RCBD designs with more than one missing value, Yate’s formula for estimating a single missing observation is used to obtain initial estimates for all missing values. These are passed to a function minimization routine to obtain the values that minimize SSE.
For other designs, specify design=2 and IMSLS_GET_SS. The function get_ss is used to obtain the Yates missing value estimates by selecting the estimates that minimize sum of squares returned by get_ss. When called, get_ss calculates the error sum of squares at each iteration assuming that the data matrix it receives is balanced and contains no missing values.
Example
Missing values can occur in any experiment. Estimating missing values via the Yates method is usually done by minimizing the error sum of squares for that experiment. If only a single observation is missing and there is an analytical formula for calculating the error sum of squares then a formula for estimating the missing value is fairly easily derived. Consider for example a split-plot experiment with a single missing value.
Suppose, for example, that 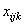 , the observation for the
ith whole-plot, jth split plot and kth block is missing.
Then the estimate for a single missing observation in the ith whole plot
is equal to:
, the observation for the
ith whole-plot, jth split plot and kth block is missing.
Then the estimate for a single missing observation in the ith whole plot
is equal to:
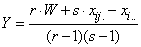 , where
, where
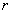 = number of blocks,
= number of blocks, 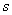 = number of split-plots,
= number of split-plots, 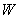 = total of all
non-missing values in same block as the missing observation,
= total of all
non-missing values in same block as the missing observation, 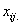 = total of the non-missing
observations across blocks of observations from ith whole-plot factor
level and the jth split-plot level, and
= total of the non-missing
observations across blocks of observations from ith whole-plot factor
level and the jth split-plot level, and 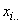 = the total of all observations,
across split-plots and blocks of the non-missing observations for the ith
whole plot.
= the total of all observations,
across split-plots and blocks of the non-missing observations for the ith
whole plot.
If more than a single observation is missing, then an iterative solution is required to obtain missing value estimates that minimize the error sum of squares.
Function imsls_f_yates simplifies this procedure. Consider, for example, a split-plot experiment conducted at a single location using fixed-effects whole and split plots. If there are no missing values, then the error sum of squares can be calculated from a 3-way analysis of variance using whole-plot, split-plot and blocks as the 3 factors. For balanced data without missing values, the errors sum of squares would be equal to the sum of the 3-way interaction between these factors and the split-plot by block interaction.
Calculating the error sum of squares using this 3-way analysis of variance is achieved using the anova_factorial routine.
float get_ss(int n, int n_independent, int *n_levels, float *x)
{
/* This routine assumes that the first three columns of dataMatrix */
/* contain the whole-plot,split-plot and block identifiers in that */
/* order. The last column of this matrix, the fourth column, must */
/* contain the observations from the experiment. It is assumed that */
/* dataMatrix is balanced and does not contain any missing */
/* observations. */
int i;
float errorSS, pValue;
float *test_effects = NULL;
float *anova_table = NULL;
float responses[24];
/* Copy responses from the last column of x into a 1-D array */
/* as expected by imsls_f_anova_factorial. */
for (i=0;i<n;i++) {
responses[i] = x[i*(n_independent+1)+n_independent];
}
/* Compute the error sum of squares. */
pValue = imsls_f_anova_factorial(n_independent, n_levels, responses,
IMSLS_TEST_EFFECTS, &test_effects,
IMSLS_ANOVA_TABLE, &anova_table,
IMSLS_POOL_INTERACTIONS, 0);
errorSS = anova_table[4] + test_effects[21];
/* Free memory returned by imsls_f_anova_factorial. */
if (test_effects != NULL) free(test_effects);
if (anova_table != NULL) free(anova_table);
return errorSS;
}
The above function is passed to the imsls_f_yates as an argument, together with a matrix containing the data for the split-plot experiment. For this example, the following data matrix obtained from an agricultural experiment will be used. In this experiment, 4 whole plots were randomly assigned to two 2 blocks. Whole-plots were subdivided into 2 split-plots. The whole-plot factor consisted of 4 different seed lots, and the split-plot factor consisted of 2 seed protectants. The data matrix of this example is a n=24 by 4 matrix with two missing observations.

The following program uses these data with imsls_f_yates to replace the two missing values with Yates estimates.
#include <stdlib.h>
#include "imsls.h"
float get_ss(int n, int n_independent, int *n_levels, float *x);
#define N 24
#define N_INDEPENDENT 3
void main()
{
char *col_labels[] = {" ", "Whole", "Split", "Block", " "};
int i;
int n = N;
int n_independent = N_INDEPENDENT;
int whole[N]={1,1,1,1,1,1,
2,2,2,2,2,2,
3,3,3,3,3,3,
4,4,4,4,4,4};
int split[N]={1,2,3,1,2,3,
1,2,3,1,2,3,
1,2,3,1,2,3,
1,2,3,1,2,3};
int block[N]={1,1,1,2,2,2,
1,1,1,2,2,2,
1,1,1,2,2,2,
1,1,1,2,2,2};
float y[N] ={0.0, 53.8, 49.5, 41.6, 0.0, 53.8,
53.3, 57.6, 59.8, 69.6, 69.6, 65.8,
62.3, 63.4, 64.5, 58.5, 50.4, 46.1,
75.4, 70.3, 68.8, 65.6, 67.3, 65.3};
float x[N][N_INDEPENDENT+1];
float error_ss;
int *missing_idx;
int n_missing;
/* Set the first and fifth observations to missing values. */
y[0] = imsls_f_machine(6);
y[4] = imsls_f_machine(6);
/* Fill the array x with the classification variables and observations. */
for (i=0;i<n; i++) {
x[i][0] = (float)whole[i];
x[i][1] = (float)split[i];
x[i][2] = (float)block[i];
x[i][3] = y[i];
}
/* Sort the data since imsls_f_anova_factorial expects sorted data. */
imsls_f_sort_data(n, n_independent+1, (float*)x, 3, 0);
n_missing = imsls_f_yates(n, n_independent, (float *)&(x[0][0]),
IMSLS_DESIGN, 2,
IMSLS_GET_SS, get_ss,
IMSLS_ERROR_SS, &error_ss,
IMSLS_MISSING_INDEX, &missing_idx,
0);
printf("Returned error sum of squares = %f\n\n", error_ss);
printf("Missing values replaced: %d\n", n_missing);
printf("Whole Split Block Estimate\n");
for (i=0;i<n_missing;i++) {
printf("%3d %3d %3d %7.3f\n",
(int)x[missing_idx[i]][0],
(int)x[missing_idx[i]][1],
(int)x[missing_idx[i]][2],
x[missing_idx[i]][n_independent]);
}
imsls_f_write_matrix("Sorted x, with estimates", n, n_independent+1,
(float*)x,
IMSLS_WRITE_FORMAT, "%-4.0f%-4.0f%-4.0f%5.2f",
IMSLS_COL_LABELS, col_labels,
IMSLS_NO_ROW_LABELS, 0);
}
float get_ss(int n, int n_independent, int *n_levels, float *x)
{
int i;
float errorSS, pValue;
float *test_effects = NULL;
float *anova_table = NULL;
float responses[24];
/*
* Copy responses from the last column of x into a 1-D array
* as expected by imsls_f_anova_factorial.
*/
for (i=0;i<n;i++) {
responses[i] = x[i*(n_independent+1)+n_independent];
}
/*
* Compute the error sum of squares.
*/
pValue = imsls_f_anova_factorial(n_independent, n_levels, responses,
IMSLS_TEST_EFFECTS, &test_effects,
IMSLS_ANOVA_TABLE, &anova_table,
IMSLS_POOL_INTERACTIONS, 0);
errorSS = anova_table[4] + test_effects[21];
/* Free memory returned by imsls_f_anova_factorial. */
if (test_effects != NULL) free(test_effects);
if (anova_table != NULL) free(anova_table);
return errorSS;
}
After running this code to replace missing values with Yates estimates, it would be followed by a call to the split-plot analysis of variance:
float *aov_table, y[24];
int expunit[24], whole[24], split[24];
for(int i=0; i < 24; i++){whole[i] = x[i]; split[i] = x[i+24];
expunit[i]= x[i+48]; y[i] = x[i+72];}
float aov_table = imsls_f_split_plot (24, 1, 4, 3, expunit, whole,
split, y[], 0);
Output
Returned error sum of squares = 95.620010
Missing values replaced: 2
Whole Split Block Estimate
1 1 1 37.300
1 2 2 58.100
Sorted x, with estimates
Whole Split Block
1 1 1 37.30
1 1 2 41.60
1 2 1 53.80
1 2 2 58.10
1 3 1 49.50
1 3 2 53.80
2 1 1 53.30
2 1 2 69.60
2 2 1 57.60
2 2 2 69.60
2 3 1 59.80
2 3 2 65.80
3 1 1 62.30
3 1 2 58.50
3 2 1 63.40
3 2 2 50.40
3 3 1 64.50
3 3 2 46.10
4 1 1 75.40
4 1 2 65.60
4 2 1 70.30
4 2 2 67.30
4 3 1 68.80
4 3 2 65.30
|
Visual Numerics, Inc. PHONE: 713.784.3131 FAX:713.781.9260 |