Performs a chi-squared goodness-of-fit test.
Synopsis
#include <imsl.h>
float imsl_f_chi_squared_test (float user_proc_cdf(), int n_observations, int n_categories, float x[], …, 0)
The type double function is imsl_d_chi_squared_test.
Required Arguments
float user_proc_cdf
(float
y) (Input)
User-supplied function that returns the
hypothesized, cumulative distribution function at the point y.
int n_observations
(Input)
The number of data elements input in x.
int n_categories
(Input)
The number of cells into which the observations are to be
tallied.
float x[]
(Input)
Array with n_observations
components containing the vector of data elements for this test.
Return Value
The p-value for the goodness-of-fit chi-squared statistic.
Synopsis with Optional Arguments
#include <imsl.h>
float imsl_f_chi_squared_test (float user_proc_cdf(), int n_observations, int n_categories, float x[],
IMSL_N_PARAMETERS_ESTIMATED, int n_parameters,
IMSL_CUTPOINTS, float **p_cutpoints,
IMSL_CUTPOINTS_USER, float cutpoints[],
IMSL_CUTPOINTS_EQUAL,
IMSL_CHI_SQUARED, float *chi_squared,
IMSL_DEGREES_OF_FREEDOM, float *df,
IMSL_FREQUENCIES, float frequencies[],
IMSL_BOUNDS, float lower_bound, float upper_bound,
IMSL_CELL_COUNTS, float **p_cell_counts,
IMSL_CELL_COUNTS_USER, float cell_counts[],
IMSL_CELL_EXPECTED, float **p_cell_expected,
IMSL_CELL_EXPECTED_USER, float cell_expected[],
IMSL_CELL_CHI_SQUARED, float **p_cell_chi_squared,
IMSL_CELL_CHI_SQUARED_USER, float cell_chi_squared[],
IMSL_FCN_W_DATA, float user_proc_cdf(), void *data,
0)
Optional Arguments
IMSL_N_PARAMETERS_ESTIMATED, int n_parameters
(Input)
The number of parameters estimated in computing the cumulative
distribution function.
IMSL_CUTPOINTS, float **p_cutpoints
(Output)
The address of a pointer to the cutpoints array. On return, the
pointer is initialized (through a memory allocation request to malloc), and the array
is stored there. Typically, float *p_cutpoints is
declared; &p_cutpoints is
used as an argument to this function; and imsl_free(p_cutpoints) is
used to free this array.
IMSL_CUTPOINTS_USER, float cutpoints[]
(Input or Output)
Array with n_categories − 1
components containing the vector of cutpoints defining the cell intervals. The
intervals defined by the cutpoints are such that the lower endpoint is not
included, and the upper endpoint is included in any interval. If IMSL_CUTPOINTS_EQUAL
is specified, equal probability cutpoints are computed and returned in cutpoints.
IMSL_CUTPOINTS_EQUAL
If
IMSL_CUTPOINTS_USER is
specified, then equal probability cutpoints can still be used if, in addition,
the IMSL_CUTPOINTS_EQUAL
option is specified. If IMSL_CUTPOINTS_USER is
not specified, equal probability cutpoints are used by default.
IMSL_CHI_SQUARED, float *chi_squared
(Output)
If specified, the chi-squared test statistic is returned in *chi_squared.
IMSL_DEGREES_OF_FREEDOM, float *df
(Output)
If specified, the degrees of freedom for the chi-squared
goodness-of-fit test is returned in *df.
IMSL_FREQUENCIES, float frequencies[]
(Input)
Array with n_observations
components containing the vector frequencies for the observations stored in
x.
IMSL_BOUNDS, float lower_bound, float upper_bound
(Input)
If IMSL_BOUNDS is
specified, then lower_bound is the
lower bound of the range of the distribution, and upper_bound is the
upper bound of this range. If lower_bound = upper_bound, a range
on the whole real line is used (the default). If the lower and upper endpoints
are different, points outside the range of these bounds are ignored.
Distributions conditional on a range can be specified when IMSL_BOUNDS is used.
By convention, lower_bound is
excluded from the first interval, but upper_bound is
included in the last interval.
IMSL_CELL_COUNTS, float **p_cell_counts
(Output)
The address of a pointer to an array containing the cell counts. The
cell counts are the observed frequencies in each of the n_categories cells. On
return, the pointer is initialized (through a memory allocation request to malloc), and the array
is stored there. Typically, float *p_cell_counts is
declared; &p_cell_counts is
used as an argument to this function; and imsl_free(p_cell_counts) is used
to free this array.
IMSL_CELL_COUNTS_USER, float cell_counts[]
(Output)
If specified, the n_categories cell
counts are returned in the array cell_counts provided
by the user.
IMSL_CELL_EXPECTED, float **p_cell_expected
(Output)
The address of a pointer to the cell expected values. The expected
value of a cell is the expected count in the cell given that the hypothesized
distribution is correct. On return, the pointer is initialized (through a memory
allocation request to malloc), and the array
is stored there. Typically, float *p_cell_expected is
declared; &p_cell_expected
is used as an argument to this function; and imsl_free(p_cell_expected) is
used to free this array.
IMSL_CELL_EXPECTED_USER, float cell_expected[]
(Output)
If specified, the n_categories cell
expected values are returned in the array cell_expected provided
by the user.
IMSL_CELL_CHI_SQUARED, float **p_cell_chi_squared
(Output)
The address of a pointer to an array of length n_categories
containing the cell contributions to chi-squared. On return, the pointer is
initialized (through a memory allocation request to malloc), and the array
is stored there. Typically, float *p_cell_chi_squared is
declared; &p_cell_chi_squared
is used as an argument to this function; and imsl_free(p_cell_chi_squared) is
used to free this array.
IMSL_CELL_CHI_SQUARED_USER, float cell_chi_squared[]
(Output)
If specified, the cell contributions to chi-squared are returned in
the array cell_chi_squared
provided by the user.
IMSL_FCN_W_DATA,
float user_proc_cdf (float y, void *data),
void *data,
(Input)
User supplied function that returns the hypothesized, cumulative
distribution function at the point y, which also accepts a pointer to
data that is supplied by the user. data is a pointer to
the data to be passed to the user-supplied function. See the Introduction, Passing Data to
User-Supplied Functions at the beginning of this manual for more
details.
Description
The function imsl_f_chi_squared_test performs a chi-squared goodness-of-fit test that a random sample of observations is distributed according to a specified theoretical cumulative distribution. The theoretical distribution, which may be continuous, discrete, or a mixture of discrete and continuous distributions, is specified via the user-defined function user_proc_cdf. Because the user is allowed to give a range for the observations, a test conditional upon the specified range is performed.
Argument n_categories gives the number of intervals into which the observations are to be divided. By default, equiprobable intervals are computed by imsl_f_chi_squared_test, but intervals that are not equiprobable can be specified (through the use of optional argument IMSL_CUTPOINTS).
Regardless of the method used to obtain the cutpoints, the intervals are such that the lower endpoint is not included in the interval, while the upper endpoint is always included. If the cumulative distribution function has discrete elements, then user-provided cutpoints should always be used since imsl_f_chi_squared_test cannot determine the discrete elements in discrete distributions.
By default, the lower and upper endpoints of the first and last intervals are − ∞ and + ∞, respectively. If IMSL_BOUNDS is specified, the endpoints are defined by the user via the two arguments lower_bound and upper_bound.
A tally of counts is maintained for the observations in x as follows. If the cutpoints are specified by the user, the tally is made in the interval to which xi belongs using the endpoints specified by the user. If the cutpoints are determined by imsl_f_chi_squared_test, then the cumulative probability at xi, F(xi), is computed via the function user_proc_cdf. The tally for xi is made in interval number
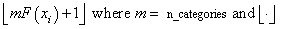
is the function that takes the greatest integer that is no larger than the argument of the function. Thus, if the computer time required to calculate the cumulative distribution function is large, user-specified cutpoints may be preferred to reduce the total computing time.
If the expected count in any cell is less than 1, then a rule of thumb is that the chi-squared approximation may be suspect. A warning message to this effect is issued in this case, as well as when an expected value is less than 5.
On some platforms, imsl_f_chi_squared_test can evaluate the user-supplied function user_proc_cdf in parallel. This is done only if the function imsl_omp_options is called to flag user-defined functions as thread-safe. A function is thread-safe if there are no dependencies between calls. Such dependencies are usually the result of writing to global or static variables
Programming Notes
The user must supply a function user_proc_cdf with calling sequence user_proc_cdf(y), that returns the value of the cumulative distribution function at any point y in the (optionally) specified range. Many of the cumulative distribution functions in Chapter 9, “Special Functions,” can be used for user_proc_cdf, either directly, if the calling sequence is correct, or indirectly, if, for example, the sample means and standard deviations are to be used in computing the theoretical cumulative distribution function.
Examples
Example 1
This example illustrates the use of imsl_f_chi_squared_test on a randomly generated sample from the normal distribution. One-thousand randomly generated observations are tallied into 10 equiprobable intervals. The null hypothesis that the sample is from a normal distribution is specified by use of the imsl_f_normal_cdf (see Chapter 9, “Special Functions”) as the hypothesized distribution function. In this example, the null hypothesis is not rejected.
#include <imsl.h>
#define SEED 123457
#define N_CATEGORIES 10
#define N_OBSERVATIONS 1000
int main()
{
float *x, p_value;
imsl_omp_options(IMSL_SET_FUNCTIONS_THREAD_SAFE, 1, 0);
imsl_random_seed_set(SEED);
/* Generate Normal deviates */
x = imsl_f_random_normal (N_OBSERVATIONS, 0);
/* Perform chi squared test */
p_value = imsl_f_chi_squared_test (imsl_f_normal_cdf, N_OBSERVATIONS,
N_CATEGORIES, x, 0);
/* Print results */
printf ("p value %7.4f\n", p_value);
}
Output
p value 0.1546
Example 2
In this example, some optional arguments are used for the data in the initial example.
#include <imsl.h>
#define SEED 123457
#define N_CATEGORIES 10
#define N_OBSERVATIONS 1000
int main()
{
float *cell_counts, *cutpoints, *cell_chi_squared;
float chi_squared_statistics[3], *x;
char *stat_row_labels[] = {"chi-squared", "degrees of freedom",
"p-value"};
imsl_omp_options(IMSL_SET_FUNCTIONS_THREAD_SAFE, 1, 0);
imsl_random_seed_set(SEED);
/* Generate Normal deviates */
x = imsl_f_random_normal (N_OBSERVATIONS, 0);
/* Perform chi squared test */
chi_squared_statistics[2] =
imsl_f_chi_squared_test (imsl_f_normal_cdf,
N_OBSERVATIONS, N_CATEGORIES, x,
IMSL_CUTPOINTS, &cutpoints,
IMSL_CELL_COUNTS, &cell_counts,
IMSL_CELL_CHI_SQUARED, &cell_chi_squared,
IMSL_CHI_SQUARED, &chi_squared_statistics[0],
IMSL_DEGREES_OF_FREEDOM, &chi_squared_statistics[1],
0);
/* Print results */
imsl_f_write_matrix ("\nChi Squared Statistics\n", 3, 1,
chi_squared_statistics,
IMSL_ROW_LABELS, stat_row_labels,
0);
imsl_f_write_matrix ("Cut Points", 1, N_CATEGORIES-1, cutpoints, 0);
imsl_f_write_matrix ("Cell Counts", 1, N_CATEGORIES, cell_counts,
0);
imsl_f_write_matrix ("Cell Contributions to Chi-Squared", 1,
N_CATEGORIES, cell_chi_squared,
0);
}
Output
Chi Squared Statistics
chi-squared 13.18
degrees of freedom 9.00
p-value 0.15
Cut Points
1 2 3 4 5 6
-1.282 -0.842 -0.524 -0.253 -0.000 0.253
7 8 9
0.524 0.842 1.282
Cell Counts
1 2 3 4 5 6
106 109 89 92 83 87
7 8 9 10
110 104 121 99
Cell Contributions to Chi-Squared
1 2 3 4 5 6
0.36 0.81 1.21 0.64 2.89 1.69
7 8 9 10
1.00 0.16 4.41 0.01
Example 3
In this example, a discrete Poisson random sample of size 1000 with parameter θ = 5.0 is generated via function imsl_f_random_poisson. In the call to imsl_f_chi_squared_test, function imsl_f_poisson_cdf is used as function user_proc_cdf.
#include <imsl.h>
#define SEED 123457
#define N_CATEGORIES 10
#define N_PARAMETERS_ESTIMATED 0
#define N_NUMBERS 1000
#define THETA 5.0
float user_proc_cdf(float);
int main()
{ int i, *poisson;
float cell_statistics[3][N_CATEGORIES];
float chi_squared_statistics[3], x[N_NUMBERS];
float cutpoints[] = {1.5, 2.5, 3.5, 4.5, 5.5, 6.5,
7.5, 8.5, 9.5};
char *cell_row_labels[] = {"count", "expected count",
"cell chi-squared"};
char *cell_col_labels[] = {"Poisson value", "0", "1", "2",
"3", "4", "5", "6", "7", "8", "9"};
char *stat_row_labels[] = {"chi-squared", "degrees of freedom",
"p-value"};
imsl_omp_options(IMSL_SET_FUNCTIONS_THREAD_SAFE, 1, 0);
imsl_random_seed_set(SEED);
/* Generate the data */
poisson = imsl_random_poisson(N_NUMBERS, THETA, 0);
/* Copy data to a floating point vector*/
for (i = 0; i < N_NUMBERS; i++)
x[i] = poisson[i];
chi_squared_statistics[2] =
imsl_f_chi_squared_test(user_proc_cdf, N_NUMBERS, N_CATEGORIES, x,
IMSL_CUTPOINTS_USER, cutpoints,
IMSL_CELL_COUNTS_USER, &cell_statistics[0][0],
IMSL_CELL_EXPECTED_USER, &cell_statistics[1][0],
IMSL_CELL_CHI_SQUARED_USER, &cell_statistics[2][0],
IMSL_CHI_SQUARED, &chi_squared_statistics[0],
IMSL_DEGREES_OF_FREEDOM, &chi_squared_statistics[1],
0);
/* Print results */
imsl_f_write_matrix("\nChi-squared statistics\n", 3, 1,
&chi_squared_statistics[0],
IMSL_ROW_LABELS, stat_row_labels,
0);
imsl_f_write_matrix("\nCell Statistics\n", 3, N_CATEGORIES,
&cell_statistics[0][0],
IMSL_ROW_LABELS, cell_row_labels,
IMSL_COL_LABELS, cell_col_labels,
0);
}
float user_proc_cdf(float k)
{
float cdf_v;
cdf_v = imsl_f_poisson_cdf ((int) k, THETA);
return cdf_v;
}
Output
Chi-squared statistics
chi-squared 10.48
degrees of freedom 9.00
p-value 0.31
Cell Statistics
Poisson value 0 1 2 3 4
count 41.0 94.0 138.0 158.0 150.0
expected count 40.4 84.2 140.4 175.5 175.5
cell chi-squared 0.0 1.1 0.0 1.7 3.7
Poisson value 5 6 7 8 9
count 159.0 116.0 75.0 37.0 32.0
expected count 146.2 104.4 65.3 36.3 31.8
cell chi-squared 1.1 1.3 1.4 0.0 0.0
Warning Errors
IMSL_EXPECTED_VAL_LESS_THAN_1 An expected value is less than 1.
IMSL_EXPECTED_VAL_LESS_THAN_5 An expected value is less than 5.
Fatal Errors
IMSL_ALL_OBSERVATIONS_MISSING All observations contain missing values.
IMSL_INCORRECT_CDF_1 The function user_proc_cdf is not a cumulative distribution function. The value at the lower bound must be nonnegative, and the value at the upper bound must not be greater than one.
IMSL_INCORRECT_CDF_2 The function user_proc_cdf is not a cumulative distribution function. The probability of the range of the distribution is not positive.
IMSL_INCORRECT_CDF_3 The function user_proc_cdf is not a cumulative distribution function. Its evaluation at an element in x is inconsistent with either the evaluation at the lower or upper bound.
IMSL_INCORRECT_CDF_4 The function user_proc_cdf is not a cumulative distribution function. Its evaluation at a cutpoint is inconsistent with either the evaluation at the lower or upper bound.
IMSL_INCORRECT_CDF_5 An error has occurred when inverting the cumulative distribution function. This function must be continuous and defined over the whole real line.
