Computes the SVD, A = USVH, of a complex rectangular matrix A. An approximate generalized inverse and rank of A also can be computed.
Synopsis
#include <imsl.h>
f_complex *imsl_c_lin_svd_gen (int m, int n, f_complex a[], …, 0)
The type d_complex function is imsl_z_lin_svd_gen.
Required Arguments
int m
(Input)
Number of rows in the matrix.
int n
(Input)
Number of columns in the matrix.
f_complex a[]
(Input)
Array of size m × n containing the
matrix.
Return Value
Using only required arguments, imsl_c_lin_svd_gen returns a pointer to a complex array of length min (m, n) containing the singular values of the matrix. To release this space, use imsl_free. If no value can be computed then NULL is returned.
Synopsis with Optional Arguments
#include <imsl.h>
f_complex *imsl_c_lin_svd_gen (int m, int n, f_complex a[],
IMSL_A_COL_DIM, int a_col_dim,
IMSL_RETURN_USER, f_complex s[],
IMSL_RANK, float tol, int *rank,
IMSL_U, f_complex **p_u,
IMSL_U_USER, f_complex u[],
IMSL_U_COL_DIM, int u_col_dim,
IMSL_V, f_complex **p_v,
IMSL_V_USER, f_complex v[],
IMSL_V_COL_DIM, int v_col_dim,
IMSL_INVERSE, f_complex **p_gen_inva,
IMSL_INVERSE_USER, f_complex gen_inva[],
IMSL_INV_COL_DIM, int gen_inva_col_dim,
0)
Optional Arguments
IMSL_A_COL_DIM, int a_col_dim
(Input)
The column dimension of the array a.
Default: a_col_dim = n
IMSL_RETURN_USER, f_complex s[]
(Output)
A user-allocated array of length min (m, n)
containing the singular values of A in its first
min (m, n) positions in nonincreasing order. The complex
entries are all real. If IMSL_RETURN_USER is
used, the return value of imsl_c_lin_svd_gen is
s.
IMSL_RANK, float tol, int *rank (Input/Output)
float tol
(Input)
Scalar containing the tolerance used to determine when a singular
value is negligible and replaced by the value zero. If tol > 0,
then a singular value si,i is considered negligible if si,i ≤ tol. If tol < 0,
then a singular value si,i is considered negligible if si,i ≤ |tol|*||A||∝. In this
case, should be an estimate of relative error or uncertainty in the data.
int *rank
(Input/Output)
Integer containing an estimate of the rank of A.
IMSL_U, f_complex **p_u
(Output)
The address of a pointer to an array of size
m × min (m, n) containing the
min (m, n) left-singular vectors of A. On return,
the necessary space is allocated by imsl_c_lin_svd_gen.
Typically, f_complex *p_u is
declared, and &p_u is used as an
argument.
IMSL_U_USER, f_complex u[]
(Output)
The address of a pointer to an array of size
m × min (m, n) containing the
min (m, n) left-singular vectors of A. If
m ≥ n, the left-singular vectors can be returned using
the storage locations of the array a.
IMSL_U_COL_DIM, int u_col_dim
(Input)
The column dimension of the array containing the left-singular
vectors.
Default: u_col_dim = min (m, n)
IMSL_V, f_complex **p_v
(Output)
The address of a pointer to an array of size
n × n containing the right singular vectors of A.
On return, the necessary space is allocated by imsl_c_lin_svd_gen.
Typically, f_complex *p_v is declared, and
&p_v is used
as an argument.
IMSL_V_USER, f_complex v[]
(Output)
The address of a pointer to an array of size
n × n containing the right singular vectors of A.
The right-singular vectors can be returned using the storage locations of the
array a. Note
that the return of the left and right-singular vectors cannot use the storage
locations of a
simultaneously.
IMSL_V_COL_DIM, int v_col_dim
(Input)
The column dimension of the array containing the right-singular
vectors.
Default: v_col_dim =
n
IMSL_INVERSE, f_complex **p_gen_inva
(Output)
The address of a pointer to an array of size
n × m containing the generalized inverse of the matrix
A. On return, the necessary space is allocated by imsl_c_lin_svd_gen.
Typically, f_complex *p_gen_inva is
declared, and &p_gen_inva is
used as an argument.
IMSL_INVERSE_USER, f_complex gen_inva[]
(Output)
A user-allocated array of size n × m
containing the general inverse of the matrix A.
IMSL_INV_COL_DIM, int gen_inva_col_dim
(Input)
The column dimension of the array containing the general inverse of
the matrix A.
Default: gen_inva_col_dim = m
Description
The function imsl_c_lin_svd_gen computes the singular value decomposition of a complex matrix A. It first reduces the matrix A to a bidiagonal matrix B by pre- and post-multiplying Householder transformations. Then, the singular value decomposition of B is computed using the implicit-shifted QR algorithm. An estimate of the rank of the matrix A is obtained by finding the smallest integer k such that sk,k ≤ tol or sk,k ≤ |tol|*||A||∝. Since si+1, ≤ si,i, it follows that all the si,i satisfy the same inequality for i = k, …, min (m, n) − 1. The rank is set to the value k − 1. If A = USVH, its generalized inverse is A+ = VS+ UT.
Here,
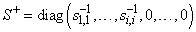
Only singular values that are not negligible are reciprocated. If IMSL_INVERSE or IMSL_INVERSE_USER is specified, the function first computes the singular value decomposition of the matrix A. The generalized inverse is then computed. The function imsl_c_lin_svd_gen fails if the QR algorithm does not converge after 30 iterations isolating an individual singular value.
Examples
Example 1
This example computes the singular values of a 6 × 3 complex matrix.
#include <imsl.h>
int main()
{
int m = 6, n = 3;
f_complex *s;
f_complex a[] = {{1.0, 2.0}, {3.0, 2.0}, {1.0,-4.0},
{3.0,-2.0}, {2.0,-4.0}, {1.0, 3.0},
{4.0, 3.0}, {-2.0,1.0}, {1.0, 4.0},
{2.0,-1.0}, {3.0, 0.0}, {3.0,-1.0},
{1.0,-5.0}, {2.0,-5.0}, {2.0, 2.0},
{1.0, 2.0}, {4.0,-2.0}, {2.0,-3.0}};
/* Compute singular values */
s = imsl_c_lin_svd_gen (m, n, a, 0);
/* Print singular values */
imsl_c_write_matrix ("Singular values", 1, n, s, 0);
}
Output
Singular values
1 2 3
( 11.77, 0.00) ( 9.30, 0.00) ( 4.99, 0.00)
Example 2
This example computes the singular value decomposition of the 6 × 3 complex matrix A. The singular values are returned in the user-provided array. The matrices U and V are returned in the space provided by the function imsl_c_lin_svd_gen.
#include <imsl.h>
int main()
{
int m = 6, n = 3;
f_complex s[3], *p_u, *p_v;
f_complex a[] = {{1.0, 2.0}, {3.0, 2.0}, {1.0,-4.0},
{3.0,-2.0}, {2.0,-4.0}, {1.0, 3.0},
{4.0, 3.0}, {-2.0,1.0}, {1.0, 4.0},
{2.0,-1.0}, {3.0, 0.0}, {3.0,-1.0},
{1.0,-5.0}, {2.0,-5.0}, {2.0, 2.0},
{1.0, 2.0}, {4.0,-2.0}, {2.0,-3.0}};
/* Compute SVD of a */
imsl_c_lin_svd_gen (m, n, a,
IMSL_RETURN_USER, s,
IMSL_U, &p_u,
IMSL_V, &p_v,
0);
/* Print decomposition factors */
imsl_c_write_matrix ("Singular values, S", 1, n, s, 0);
imsl_c_write_matrix ("Left singular vectors, U", m, n, p_u, 0);
imsl_c_write_matrix ("Right singular vectors, V", n, n, p_v, 0);
}
Output
Singular values, S
1 2 3
( 11.77, 0.00) ( 9.30, 0.00) ( 4.99, 0.00)
Left singular vectors, U
1 2 3
1 ( 0.1968, 0.2186) ( 0.5011, 0.0217) ( -0.2007, -0.1003)
2 ( 0.3443, -0.3542) ( -0.2933, 0.0248) ( 0.1155, -0.2338)
3 ( 0.1457, 0.2307) ( -0.5424, 0.1381) ( -0.4361, -0.4407)
4 ( 0.3016, -0.0844) ( 0.2157, 0.2659) ( -0.0523, -0.0894)
5 ( 0.2283, -0.6008) ( -0.1325, 0.1433) ( 0.3152, -0.0090)
6 ( 0.2876, -0.0350) ( 0.4377, -0.0400) ( 0.0458, -0.6205)
Right singular vectors, V
1 2 3
1 ( 0.6616, 0.0000) ( -0.2651, 0.0000) ( -0.7014, 0.0000)
2 ( 0.7355, 0.0379) ( 0.3850, -0.0707) ( 0.5482, 0.0624)
3 ( 0.0507, -0.1317) ( 0.1724, 0.8642) ( -0.0173, -0.4509)
Example 3
This example computes the rank and generalized inverse of a 6 × 4 matrix A. The rank and the 4 × 6 generalized inverse matrix A+ are printed.
#include <imsl.h>
int main()
{
int m = 6, n = 4;
int *rank;
float tol;
f_complex gen_inv[24], *s;
f_complex a[] = {{1.0, 2.0}, {3.0, 2.0}, {1.0,-4.0}, {1.0,0.0},
{3.0,-2.0}, {2.0,-4.0}, {1.0, 3.0}, {0.0,1.0},
{4.0, 3.0}, {-2.0,1.0}, {1.0, 4.0}, {0.0,0.0},
{2.0,-1.0}, {3.0, 0.0}, {3.0,-1.0}, {2.0,1.0},
{1.0,-5.0}, {2.0,-5.0}, {2.0, 2.0}, {1.0,3.1},
{1.0, 2.0}, {4.0,-2.0}, {2.0,-3.0}, {1.4,1.9}};
/* Factor a */
tol = 1.e-4;
s = imsl_c_lin_svd_gen (m, n, a,
IMSL_RANK, tol, &rank,
IMSL_INVERSE_USER, gen_inv,
IMSL_INV_COL_DIM, m,
0);
/* Print rank and generalized */
/* inverse matrix */
printf ("Rank = %2d", rank);
imsl_c_write_matrix ("Singular values", 1, n, s, 0);
imsl_c_write_matrix ("Generalized inverse", n, m, gen_inv,
IMSL_A_COL_DIM, m, 0);
}
Output
Rank = 4
Singular values
1 2 3
( 12.13, 0.00) ( 9.53, 0.00) ( 5.67, 0.00)
4
( 1.74, 0.00)
Generalized inverse
1 2 3
1 ( 0.0266, 0.0164) ( -0.0185, 0.0453) ( 0.0720, 0.0700)
2 ( 0.0061, 0.0280) ( 0.0820, -0.1156) ( -0.0410, -0.0242)
3 ( -0.0019, -0.0572) ( 0.1174, 0.0812) ( 0.0499, 0.0463)
4 ( 0.0380, 0.0298) ( -0.0758, -0.2158) ( 0.0356, -0.0557)
4 5 6
1 ( -0.0220, -0.0428) ( -0.0003, -0.0709) ( 0.0254, 0.1050)
2 ( 0.0959, 0.0885) ( -0.0187, 0.0287) ( -0.0218, -0.1109)
3 ( -0.0234, 0.1033) ( -0.0769, 0.0103) ( 0.0810, -0.1074)
4 ( 0.2918, -0.0763) ( 0.0881, 0.2070) ( -0.1531, 0.0814)
Warning Errors
IMSL_SLOWCONVERGENT_MATRIX Convergence cannot be reached after 30 iterations.

