Computes a matrix of dissimilarities (or similarities) between the columns (or rows) of a matrix.
Synopsis
#include <imsls.h>
float *imsls_f_dissimilarities (int nrow, int ncol, float *x, …, 0)
The type double function is imsls_d_dissimilarities.
Required Arguments
int nrow
(Input)
Number of rows in the matrix.
int ncol
(Input)
Number of columns in the matrix.
float *x (Input)
Array of size nrow by ncol containing the
matrix.
Return Value
An array of size m by m containing the computed dissimilarities or similarities, where m = nrow if optional argument IMSLS_ROWS is used, and m = ncol otherwise.
Synopsis with Optional Arguments
#include <imsls.h>
float
*imsls_f_dissimilarities
(int nrow, int
ncol,
float
*x,
IMSLS_ROWS, or
IMSLS_COLUMNS,
IMSLS_INDEX,
int
ndstm, int
ind[],
IMSLS_METHOD, int
imeth,
IMSLS_SCALE, int
iscale,
IMSLS_X_COL_DIM,
int x_col_dim,
IMSLS_RETURN_USER,
float
dist[],
0)
Optional Arguments
IMSLS_ROWS,
or
IMSLS_COLUMNS,
(Input)
Exactly one of these options can be present to indicate whether
distances are computed between rows or columns of x.
Default:
Distances are computed between rows.
IMSLS_INDEX, int
ndstm,
int ind[],
(Input)
Argument ind is an array of
length ndstm
containing the indices of the rows (columns if IMSLS_ROWS is used) to
be used in computing the distance measure.
Default: All rows(columns)
are used.
IMSLS_METHOD,
int imeth
(Input)
Method to be used in computing the dissimilarities or
similarities.
Default: imeth = 0.
|
imeth |
Method |
|
0 |
Euclidean distance (L2 norm) |
|
1 |
Sum of the absolute differences (L1 norm) |
|
2 |
Maximum difference (L∞ norm) |
|
3 |
Mahalanobis distance |
|
4 |
Absolute value of the cosine of the angle between the vectors |
|
5 |
Angle in radians (0, π) between the lines through the origin defined by the vectors |
|
6 |
Correlation coefficient |
|
7 |
Absolute value of the correlation coefficient |
|
8 |
Number of exact matches |
See the Description section for a more detailed description of each measure.
IMSLS_SCALE,
int iscale (Input)
Scaling
option. (Input)
iscale is not used for
methods 3 through 8.
Default: iscale = 0.
|
iscale |
Scaling Performed |
|
0 |
No scaling is performed. |
|
1 |
Scale each column (row, if IMSLS_ROWS is used) by the standard deviation of the column (row). |
|
2 |
Scale each column (row, if IMSLS_ROWS is used) by the range of the column (row). |
IMSLS_X_COL_DIM,
int x_col_dim
(Input)
Column dimension of x.
Default: x_col_dim = ncol.
IMSLS_RETURN_USER,
float dist[]
(Output)
User allocated array of size m by m
containing the computed dissimilarities or similarities, where m =
nrow if IMSLS_ROWS is used,
and m = ncol otherwise.
Description
Function imsls_f_dissimilarities computes an upper triangular matrix (excluding the diagonal) of dissimilarities (or similarities) between the columns or rows of a matrix. Nine different distance measures can be computed. For the first three measures, three different scaling options can be employed. Output from imsls_f_dissimilarities is generally used as input to clustering or multidimensional scaling functions.
The following discussion assumes that the distance measure is being computed between the columns of the matrix, i.e., that IMSLS_COLUMNS is used. If distances between the rows of the matrix are desired, use optional argument IMSLS_ROWS.
For imeth = 0 to 2, each row of x is first scaled according to the value of iscale. The scaling parameters are obtained from the values in the row scaled as either the standard deviation of the row or the row range; the standard deviation is computed from the unbiased estimate of the variance. If iscale is 0, no scaling is performed, and the parameters in the following discussion are all 1.0. Once the scaling value (if any) has been computed, the distance between column i and column j is computed via the difference vector zk = (xk − yk)/sk, i = 1, …, ndstm, where xk denotes the k-th element in the i-th column, and yk denotes the corresponding element in the j-th column. For given zi, the metrics 0 to 2 are defined as:
|
imeth |
Metric |
|
0 |
|
|
1 |
|
|
|
|
Distance measures corresponding to imeth = 3 to 8 do not allow for scaling. These measures are defined via the column vectors X = (xi), Y = (yi), and Z = (xi − yi) as follows:
|
imeth |
Scaling Performed |
|
|
|
|
|
|
|
5 |
θ, where θ is defined in 4. |
|
6 |
ρ = the usual (centered) estimate of the correlation between X and Y. |
|
7 |
The absolute value of ρ (where ρ is defined in 6). |
|
8 |
The number of times xi = yi, where xi and yi are elements of X and Y. |
For the Mahalanobis distance, any variable used in computing the distance measure that is (numerically) linearly dependent upon the previous variables in the ind vector is omitted from the distance measure.
Example
The following example illustrates the use of imsls_f_dissimilarities for computing the Euclidean distance between the rows of a matrix.
#include <imsls.h>
int main()
{
int ncol=2, nrow = 4;
float x [4][2] = {1., 1.,
1., 0.,
1.,-1.,
1., 2.};
float *dist;
dist = imsls_f_dissimilarities(nrow, ncol, (float*)x, 0);
imsls_f_write_matrix("dist", 4, 4, dist, 0);
}
Output
dist
1 2 3 4
1 0 1 2 1
2 0 0 1 2
3 0 0 0 3
4 0 0 0 0

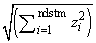
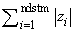
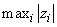
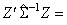
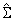
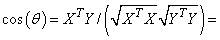 the dot product of X and Y
divided by the length of X times the length of Y .
the dot product of X and Y
divided by the length of X times the length of Y .