Performs a Kolmogorov-Smirnov one-sample test for continuous distributions.
Synopsis
#include <imsls.h>
float *imsls_f_kolmogorov_one (float cdf(), int n_observations, float x[], ..., 0)
The type double function is imsls_d_kolmogorov_one.
Required Arguments
float cdf (float
x)
(Input)
User-supplied function to compute the cumulative distribution
function (CDF)
at a given value. The form is CDF(x), where x is the value at
which cdf is to
be evaluated (Input) and cdf is the value of
CDF at x. (Output)
int
n_observations (Input)
Number of
observations.
float x[]
(Input)
Array of size n_observations
containing the observations.
Return Value
Pointer to an array of length 3 containing Z, p1, and p2 .
Synopsis with Optional Arguments
#include <imsls.h>
float
*imsls_f_kolmogorov_one (float
cdf(),
int n_observations,
float x[],
IMSLS_DIFFERENCES,
float
**differences,
IMSLS_DIFFERENCES_USER,float
differences[]
IMSLS_N_MISSING,
int
*n_missing,
IMSLS_RETURN_USER,
float
test_statistic[],
IMSLS_FCN_W_DATA,
float
cdf (),
void
*data,
0)
Optional Arguments
IMSLS_DIFFERENCES, float
**differences (Output)
Address of a pointer to the
internally allocated array containing Dn , Dn+,
Dn-.
IMSLS_DIFFERENCES_USER, float
differences[]
Storage for the array differences is
provided by the user.
See
IMSLS_DIFFERENCES.
IMSLS_N_MISSING, int
*n_missing (Ouput)
Number of missing values is returned in
*n_missing.
IMSLS_RETURN_USER, float
test_statistics[] (Output)
If specified, the
Z-score and the p-values for hypothesis test against both
one-sided and two-sided alternatives is stored in array test_statistics
provided by the user.
IMSLS_FCN_W_DATA, float cdf (float
x) ,
void
*data, (Input)
User-supplied function to compute the cumulative
distribution function, which also accepts a pointer to data that is supplied by
the user. data is a pointer to
the data to be passed to the user-supplied function. See the Introduction, Passing Data to
User-Supplied Functions at the beginning of this manual for more
details.
Description
The routine imsls_f_kolmogorov_one performs a Kolmogorov-Smirnov goodness-of-fit test in one sample. The hypotheses tested follow:
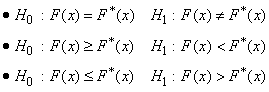
where F is the cumulative distribution function (CDF) of the random variable, and the theoretical cdf, F* , is specified via the user-supplied function cdf. Let n = n_observations − n_missing. The test statistics for both one-sided alternatives
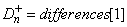
and
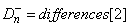
and the two-sided (Dn = differences[0])
alternative are computed as well as an asymptotic z-score (test_statistics[0])
and p-values associated with the one-sided (test_statistics[1])
and two-sided (test_statistics[2])
hypotheses. For n > 80, asymptotic p-values are used (see
Gibbons 1971). For n ≤ 80, exact one-sided p-values are computed
according to a method given by Conover (1980, page 350). An approximate
two-sided test p-value is obtained as twice the one-sided p-value.
The approximation is very close for one-sided
p-values less than 0.10
and becomes very bad as the one-sided p-values get larger.
Programming Notes
1. The theoretical CDF is assumed to be continuous. If the CDF is not continuous, the statistics
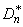
will not be computed correctly.
2. Estimation of parameters in the theoretical CDF from the sample data will tend to make the p-values associated with the test statistics too liberal. The empirical CDF will tend to be closer to the theoretical CDF than it should be.
3. No attempt is made to check that all points in the sample are in the support of the theoretical CDF. If all sample points are not in the support of the CDF, the null hypothesis must be rejected.
Example
In this example, a random sample of size 100 is generated via routine imsls_f_random_uniform (Chapter 12, “Random Number Generation”) for the uniform (0, 1) distribution. We want to test the null hypothesis that the cdf is the standard normal distribution with a mean of 0.5 and a variance equal to the uniform (0, 1) variance (1/12).
#include <imsls.h>
#include <stdio.h>
float cdf(float);
int main()
{
float *statistics=NULL, *diffs = NULL, *x=NULL;
int nobs = 100, nmiss;
imsls_random_seed_set(123457);
x = imsls_f_random_uniform(nobs, 0);
statistics = imsls_f_kolmogorov_one(cdf, nobs, x,
IMSLS_N_MISSING, &nmiss,
IMSLS_DIFFERENCES, &diffs,
0);
printf("D = %8.4f\n", diffs[0]);
printf("D+ = %8.4f\n", diffs[1]);
printf("D- = %8.4f\n", diffs[2]);
printf("Z = %8.4f\n", statistics[0]);
printf("Prob greater D one sided = %8.4f\n", statistics[1]);
printf("Prob greater D two sided = %8.4f\n", statistics[2]);
printf("N missing = %d\n", nmiss);
}
float cdf(float x)
{
float mean = .5, std = .2886751, z;
z = (x-mean)/std;
return(imsls_f_normal_cdf(z));
}
Output
D = 0.1471
D+ = 0.0810
D- = 0.1471
Z = 1.4708
Prob greater D one-sided = 0.0132
Prob greater D two-sided = 0.0264
N missing = 0
Warning Errors
IMSLS_TIE_DETECTED # ties were detected in the sample.
