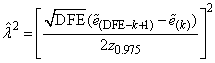Performs partial least squares (PLS) regression for one or more response variables and one or more predictor variables.
Synopsis
#include <imsls.h>
float *imsls_f_pls_regression (int ny, int h, float y[], int nx, int p, float x[], ..., 0)
The type double function is imsls_d_pls_regression.
Required Arguments
int ny
(Input)
The number of rows of y.
int h
(Input)
The number of response variables.
float y[]
(Input)
Array of length ny × h containing the
values of the responses.
int nx
(Input)
The number of rows of x.
int p
(Input)
The number of predictor variables.
float x[]
(Input)
Array of length nx × p containing
containing the values of the predictor variables.
Return Value
A pointer to the array of length ix × iy containing the final PLS regression coefficient estimates, where ix ≤ p is the number of predictor variables in the model, and iy ≤ h is the number of response variables. To release this space, use imsls_free. If the estimates cannot be computed, NULL is returned.
Synopsis with Optional Arguments
#include <imsls.h>
float
*imsls_f_pls_regression
(int ny,
int h,
float y[],
int nx,
int p,
float x[],
IMSLS_N_OBSERVATIONS,
int
nobs,
IMSLS_Y_INDICES, int
iy,
int iyind[],
IMSLS_X_INDICES, int
ix,
int ixind[],
IMSLS_N_COMPONENTS,
int
ncomps,
IMSLS_CROSS_VALIDATATION,
int
cv,
IMSLS_N_FOLD,
int
k,
IMSLS_SCALE,
int
scale,
IMSLS_PRINT_LEVEL,
int
iprint,
IMSLS_PREDICTED,
float
**yhat,
IMSLS_PREDICTED_USER,
float
yhat[],
IMSLS_RESIDUALS,
float
**resids,
IMSLS_RESIDUALS_USER,
float
resids[],
IMSLS_STD_ERRORS,
float
**se,
IMSLS_STD_ERRORS_USER,
float
se[],
IMSLS_PRESS,
float
**press,
IMSLS_PRESS_USER,
float
press[],
IMSLS_X_SCORES,
float
**xscrs,
IMSLS_X_SCORES_USER,
float
xscrs[],
IMSLS_Y_SCORES,
float
**yscrs,
IMSLS_Y_SCORES_USER,
float
yscrs[],
IMSLS_X_LOADINGS,
float
**xldgs,
IMSLS_X_LOADINGS_USER,
float
xldgs[],
IMSLS_Y_LOADINGS,
float
**yldgs,
IMSLS_Y_LOADINGS_USER,
float
yldgs[],
IMSLS_WEIGHTS,
float
**wts,
IMSLS_WEIGHTS_USER,
float
wts[],
IMSLS_RETURN_USER,
float
coef[],
0)
Optional Arguments
IMSLS_N_OBSERVATIONS,
int
nobs
(Input)
Positive integer specifying the number of observations to be
used in the analysis.
Default: nobs =
min(ny,
nx).
IMSLS_Y_INDICES,
int
iy,
int
iyind[]
(Input)
Argument iyind is an array of
length iy
containing column indices of y specifying which
response variables to use in the analysis. Each element in iyind must be less
than or equal to h-1.
Default:
iyind = 0, 1, …,
h-1.
IMSLS_X_INDICES,
int
ix,
int
ixind[]
(Input)
Argument ixind is an array of
length ix
containing column indices of x specifying
which predictor variables to use in the analysis. Each element in ixind must be less
than or equal to p-1.
Default:
ixind = 0, 1, …,
p-1.
IMSLS_N_COMPONENTS,
int ncomps
(Input)
The number of PLS components to fit. ncomps ≤ ix.
Default:
ncomps = ix.
Note: If
cv = 1 is used,
models with 1 up to ncomps components are
tested using cross-validation. The model with the lowest predicted residual sum
of squares is reported.
IMSLS_CROSS_VALIDATION,
int cv
(Input)
If cv = 0, the function
fits only the model specified by ncomps. If cv = 1, the function
performs K-fold cross validation to select the number of
components.
Default: cv = 1.
IMSLS_N_FOLD, int
k
(Input)
The number of folds to use in K-fold cross validation.
k must be
between 1 and nobs, inclusive. k is ignored if cv = 0 is
used.
Default: k = 5.
Note:
If nobs/k ≤ 3, the routine
performs leave-one-out cross validation as opposed to K-fold cross
validation.
IMSLS_SCALE, int
scale
(Input)
If scale = 1, y and x are centered
and scaled to have mean 0 and standard deviation of 1. If scale = 0, y and x are centered
to have mean 0 but are not scaled.
Default: scale = 0.
IMSLS_PRINT_LEVEL,
int iprint
(Input)
Printing option.
|
iprint |
Action |
|
0 |
No Printing. |
|
1 |
Prints final results only. |
|
2 |
Prints intermediate and final results. |
Default: iprint = 0.
IMSLS_PREDICTED,
float **yhat
(Ouput)
Argument yhat is the address of
an array of length nobs × iy, containing the
predicted values for the response variables using the final values of the
coefficients.
IMSLS_PREDICTED_USER,
float yhat[]
(Ouput)
Storage for array yhat is provided by
the user. See IMSLS_PREDICTED.
IMSLS_RESIDUALS,
float **resids
(Ouput)
Argument resids is the address
of an array of length nobs × iy, containing
residuals of the final fit for each response variable.
IMSLS_RESIDUALS_USER,
float resids[]
(Ouput)
Storage for array resids is provided by
the user. See IMSLS_RESIDUALS.
IMSLS_STD_ERRORS,
float **se
(Ouput)
Argument se is the address of
an array of length ix × iy, containing the
standard errors of the PLS coefficients.
IMSLS_STD_ERRORS_USER,
float se[]
(Ouput)
Storage for array se is provided by the
user. See IMSLS_STD_ERRORS.
IMSLS_PRESS, float
**press
(Ouput)
Argument press is the address
of an array of length ncomps × iy, containing the
predicted residual error sum of squares obtained by cross-validation for each
model of size j = 1, … , ncomps
components. The argument press is ignored if
cv = 0
is used for IMSLS_CROSS_VALIDATION.
IMSLS_PRESS_USER,
float press[]
(Ouput)
Storage for array press is provided by
the user. See IMSLS_PRESS.
IMSLS_X_SCORES,
float **xscrs
(Ouput)
Argument xscrs is the address
of an array of length nobs × ncomps
containing X-scores.
IMSLS_X_SCORES_USER,
float xscrs[]
(Ouput)
Storage for array xscrs is provided by
the user. See IMSLS_X_SCORES.
IMSLS_Y_SCORES,
float **yscrs
(Ouput)
Argument yscrs is the address
of an array of length nobs × ncomps containing
Y-scores.
IMSLS_Y_SCORES_USER,
float yscrs[]
(Ouput)
Storage for array yscrs is provided by
the user. See IMSLS_Y_SCORES.
IMSLS_X_LOADINGS,
float **xldgs
(Ouput)
Argument xldgs is the address
of an array of length ix × ncomps, containing
X-loadings.
IMSLS_X_LOADINGS_USER,
float xldgs[]
(Ouput)
Storage for array xldgs is provided by
the user. See IMSLS_X_LOADINGS.
IMSLS_Y_LOADINGS,
float **yldgs
(Ouput)
Argument yldgs is the address
of an array of length iy × ncomps, containing
Y-loadings.
IMSLS_Y_LOADINGS_USER,
float yldgs[]
(Ouput)
Storage for array yldgs is provided by
the user. See IMSLS_Y_LOADINGS.
IMSLS_WEIGHTS,
float **wts
(Ouput)
Argument wts is the address of
an array of length ix × ncomps, containing the
weight vectors.
IMSLS_WEIGHTS_USER,
float wts[]
(Ouput)
Storage for array wts is provided by the
user. See IMSLS_WEIGHTS.
IMSLS_RETURN_USER,
float coef[]
(Ouput)
If specified, the final PLS regression coefficient estimates
are stored in array coef provided by the
user.
Description
Function imsls_f_pls_regression
performs partial least squares regression for a response matrix. 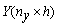 and a set of p
explanatory variables,
and a set of p
explanatory variables, 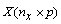 . imsls_f_pls_regression
finds linear combinations of the predictor variables that have highest
covariance with Y. In so doing, imsls_f_pls_regression
produces a predictive model for Y using components (linear combinations)
of the individual predictors. Other names for these linear combinations
are scores, factors, or latent variables. Partial least squares regression
is an alternative method to ordinary least squares for problems with many,
highly collinear predictor variables. For further discussion see, for
example, Abdi (2009), and Frank and Friedman (1993).
. imsls_f_pls_regression
finds linear combinations of the predictor variables that have highest
covariance with Y. In so doing, imsls_f_pls_regression
produces a predictive model for Y using components (linear combinations)
of the individual predictors. Other names for these linear combinations
are scores, factors, or latent variables. Partial least squares regression
is an alternative method to ordinary least squares for problems with many,
highly collinear predictor variables. For further discussion see, for
example, Abdi (2009), and Frank and Friedman (1993).
In Partial Least Squares (PLS), a score, or component matrix, T, is selected to represent both X and Y as in,
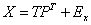
and
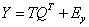
The matrices P and Q are the least squares solutions of X and Y regressed on T.
That is,
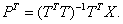
and
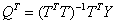
The columns of T in the above relations are often called X-scores, while the columns of P are the X-loadings. The columns of the matrix U in Y = UQT + G are the corresponding Y scores, where G is a residual matrix and Q, as defined above, contains the Y-loadings.
Restricting T to be linear in X , the problem is to find a set of weight vectors (columns of W) such that T = XW predicts both X and Y reasonably well.
Formally, 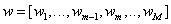 where each wj is a column vector of
length p, M ≤ p is the number of components, and
where the m-th partial least squares (PLS) component wm solves:
where each wj is a column vector of
length p, M ≤ p is the number of components, and
where the m-th partial least squares (PLS) component wm solves:
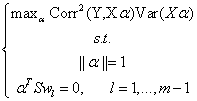
where 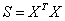 and
and 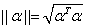 is the Euclidean norm. For
further details see Hastie, et. al., pages 80-82 (2001).
is the Euclidean norm. For
further details see Hastie, et. al., pages 80-82 (2001).
That is, wm is the vector which
maximizes the product of the squared correlation between Y and Xα
and the variance of Xα, subject to being orthogonal to each previous
weight vector left multiplied by S. The PLS regression coefficients 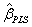 arise from
arise from
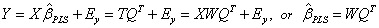
Algorithms to solve the above optimization problem include NIPALS (nonlinear iterative partial least squares) developed by Herman Wold (1966, 1985) and numerous variations, including the SIMPLS algorithm of de Jong (1993). imsls_f_pls_regression implements the SIMPLS method. SIMPLS is appealing because it finds a solution in terms of the original predictor variables, whereas NIPALS reduces the matrices at each step. For univariate Y it has been shown that SIMPLS and NIPALS are equivalent (the score, loading, and weights matrices will be proportional between the two methods).
By default, imsls_f_pls_regression searches for the best number of PLS components using K-fold cross-validation. That is, for each M = 1, 2,…, p, imsls_f_pls_regression estimates a PLS model with M components using all of the data except a hold-out set of size roughly equal to nobs/k. Using the resulting model estimates, imsls_f_pls_regression predicts the outcomes in the hold-out set and calculates the predicted residual sum of squares (PRESS). The procedure then selects the next hold-out sample and repeats for a total of K times (i.e., folds). For further details see Hastie, et. al., pages 241-245 (2001).
For each response variable, imsls_f_pls_regression returns results for the model with lowest PRESS. The best model (the number of components giving lowest PRESS), generally will be different for different response variables.
When requested via the optional argument, IMSLS_STD_ERRORS, imsls_f_pls_regression calculates modifed jackknife estimates of the standard errors as described in Martens and Martens (2000).
Comments
1. imsls_f_pls_regression defaults to leave-one-out cross-validation when there are too few observations to form K folds in the data. The user is cautioned that there may be too few observations to make strong inferences from the results.
2. This implementation of imsls_f_pls_regression does not handle missing values. The user should remove missing values or NaN’s from the input data.
Examples
Example 1
The following artificial data set is provided in de Jong (1993).
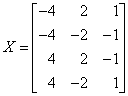
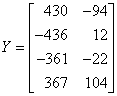
The first call to imsls_f_pls_regression fixes the number of components to 3 for both response variables, and the second call performs K-fold cross validation. Note that because n is small, imsls_f_pls_regression performs leave-one-out (LOO) cross–validation.
#include <imsls.h>
#include <stdio.h>
#define H 2
#define N 4
#define P 3
int main() {
int iprint=1, ncomps=3;
float x[N][P] = {
-4.0, 2.0, 1.0,
-4.0, -2.0, -1.0,
4.0, 2.0, -1.0,
4.0, -2.0, 1.0
};
float y[N][H] = {
430.0, -94.0,
-436.0, 12.0,
-361.0, -22.0,
367.0, 104.0
};
float *coef=NULL, *yhat=NULL, *se=NULL;
float *coef2=NULL, *yhat2=NULL, *se2=NULL;
/* Print out informational error. */
imsls_error_options(IMSLS_SET_PRINT, IMSLS_ALERT, 1, 0);
printf("Example 1a: no cross-validation, request %d components.\n",
ncomps);
coef = imsls_f_pls_regression(N, H, &y[0][0], N, P, &x[0][0],
IMSLS_N_COMPONENTS, ncomps,
IMSLS_CROSS_VALIDATION, 0,
IMSLS_PRINT_LEVEL, iprint,
IMSLS_PREDICTED, &yhat,
IMSLS_STD_ERRORS, &se,
0);
printf("\nExample 1b: cross-validation\n");
coef2 = imsls_f_pls_regression(N, H, &y[0][0], N, P, &x[0][0],
IMSLS_PRINT_LEVEL, iprint,
IMSLS_PREDICTED, &yhat2,
IMSLS_STD_ERRORS, &se2,
0);
}
Output
Example 1a: no cross-validation, request 3 components.
PLS Coeff
1 2
1 0.7 10.2
2 17.3 -29.0
3 398.5 5.0
Predicted Y
1 2
1 430 -94
2 -436 12
3 -361 -22
4 367 104
Std. Errors
1 2
1 131.5 5.1
2 263.0 10.3
3 526.0 20.5
*** ALERT Error IMSLS_RESIDUAL_CONVERGED from imsls_f_pls_regression.
*** For response 2, residuals converged in 2 components, while 3 is
*** the requested number of components.
Example 1b: cross-validation
Cross-validated results for response 1:
Comp PRESS
1 542904
2 830050
3 830050
The best model has 1 component(s).
Cross-validated results for response 2:
Comp PRESS
1 5080
2 1263
3 1263
The best modesl has 2 component(s).
PLS Coeff
1 2
1 15.9 12.7
2 49.2 -23.9
3 371.1 0.6
Predicted Y
1 2
1 405.8 -97.8
2 -533.3 -3.5
3 -208.8 2.2
4 336.4 99.1
Std. Errors
1 2
1 134.1 7.1
2 269.9 3.8
3 478.5 19.5
*** ALERT Error IMSLS_RESIDUAL_CONVERGED from imsls_f_pls_regression.
*** For response 2, residuals converged in 2 components, while 3 is
*** the requested number of components.
Example 2
The data, as appears in S. Wold, et.al. (2001), is a single response variable, the “free energy of the unfolding of a protein”, while the predictor variables are 7 different, highly correlated measurements taken on 19 amino acids.
#include <imsls.h>
#include <stdio.h>
#define H 1
#define N 19
#define P 7
int main() {
int iprint=2, ncomps=7;
float x[N][P] = {
0.23, 0.31, -0.55, 254.2, 2.126, -0.02, 82.2,
-0.48, -0.6, 0.51, 303.6, 2.994, -1.24, 112.3,
-0.61, -0.77, 1.2, 287.9, 2.994, -1.08, 103.7,
0.45, 1.54, -1.4, 282.9, 2.933, -0.11, 99.1,
-0.11, -0.22, 0.29, 335.0, 3.458, -1.19, 127.5,
-0.51, -0.64, 0.76, 311.6, 3.243, -1.43, 120.5,
0.0, 0.0, 0.0, 224.9, 1.662, 0.03, 65.0,
0.15, 0.13, -0.25, 337.2, 3.856, -1.06, 140.6,
1.2, 1.8, -2.1, 322.6, 3.35, 0.04, 131.7,
1.28, 1.7, -2.0, 324.0, 3.518, 0.12, 131.5,
-0.77, -0.99, 0.78, 336.6, 2.933, -2.26, 144.3,
0.9, 1.23, -1.6, 336.3, 3.86, -0.33, 132.3,
1.56, 1.79, -2.6, 366.1, 4.638, -0.05, 155.8,
0.38, 0.49, -1.5, 288.5, 2.876, -0.31, 106.7,
0.0, -0.04, 0.09, 266.7, 2.279, -0.4, 88.5,
0.17, 0.26, -0.58, 283.9, 2.743, -0.53, 105.3,
1.85, 2.25, -2.7, 401.8, 5.755, -0.31, 185.9,
0.89, 0.96, -1.7, 377.8, 4.791, -0.84, 162.7,
0.71, 1.22, -1.6, 295.1, 3.054, -0.13, 115.6
};
float y[N][H] = {8.5, 8.2, 8.5, 11.0, 6.3, 8.8, 7.1, 10.1,
16.8, 15.0, 7.9, 13.3, 11.2, 8.2, 7.4, 8.8, 9.9, 8.8, 12.0};
float *coef=NULL, *yhat=NULL, *se=NULL;
float *coef2=NULL, *yhat2=NULL, *se2=NULL;
printf("Example 2a: no cross-validation, request %d components.\n",
ncomps);
coef = imsls_f_pls_regression(N, H, &y[0][0], N, P, &x[0][0],
IMSLS_N_COMPONENTS, ncomps,
IMSLS_CROSS_VALIDATION, 0,
IMSLS_SCALE, 1,
IMSLS_PRINT_LEVEL, iprint,
IMSLS_PREDICTED, &yhat,
IMSLS_STD_ERRORS, &se,
0);
printf("\nExample 2b: cross-validation\n");
coef2 = imsls_f_pls_regression(N, H, &y[0][0], N, P, &x[0][0],
IMSLS_SCALE, 1,
IMSLS_PRINT_LEVEL, iprint,
IMSLS_PREDICTED, &yhat2,
IMSLS_STD_ERRORS, &se2,
0);
}
Output
Example 2a: no cross-validation, request 7 components.
Standard PLS Coefficients
1
-5.455
1.668
0.625
1.433
-2.550
4.858
4.852
PLS Coeff
1
-20.02
4.63
1.42
0.09
-7.27
20.88
0.46
Predicted Y
1
9.38
7.30
8.09
12.02
8.80
6.76
7.24
10.44
15.79
14.35
8.42
9.95
11.52
8.64
8.23
8.40
11.12
8.97
12.39
Std. Errors
1
3.513
2.423
0.801
3.190
1.627
3.272
3.518
Corrected Std. Errors
1
12.89
6.72
1.82
0.20
4.64
14.06
0.33
Variance Analysis
=============================================
Pctge of Y variance explained
Component Cum. Pctge
1 39.7
2 42.8
3 58.3
4 65.1
5 68.1
6 75.0
7 75.5
=============================================
Pctge of X variance explained
Component Cum. Pctge
1 64.1
2 96.3
3 97.4
4 97.9
5 98.2
6 98.3
7 98.4
Example 2b: cross-validation
Cross-validated results for response 1:
Comp PRESS
1 0.642
2 0.663
3 0.937
4 1.058
5 2.224
6 1.581
7 1.206
The best model has 1 component(s).
Standard PLS Coefficients
1
0.1486
0.1639
-0.1492
0.0617
0.0669
0.1150
0.0691
PLS Coeff
1
0.5453
0.4546
-0.3384
0.0039
0.1907
0.4942
0.0065
Predicted Y
1
9.18
7.97
7.55
10.48
8.75
7.89
8.44
9.47
11.76
11.80
7.37
11.14
12.65
9.96
8.61
9.27
13.47
11.33
10.71
Std. Errors
1
0.06312
0.07055
0.06272
0.03911
0.03364
0.06386
0.03955
Corrected Std. Errors
1
0.2317
0.1957
0.1423
0.0025
0.0959
0.2745
0.0037
Variance Analysis
=============================================
Pctge of Y variance explained
Component Cum. Pctge
1 39.7
=============================================
Pctge of X variance explained
Component Cum. Pctge
1 64.1
Alert Errors
IMSLS_RESIDUAL_CONVERGED For response #, residuals converged in # components, while #s is the requested number of components.