Example 3: Three-way Analysis of Variance
This example performs a three-way analysis of variance using data discussed by John (1971, pp. 91 92). The responses are weights (in grams) of roots of carrots grown with varying amounts of applied nitrogen (
A), potassium (
B), and phosphorus (
C). Each cell of the three-way layout has one response. Note that the ABC interactions sum of squares, which is 186, is given incorrectly by John (1971, Table 5.2.) The three-way layout is given in the following table:
| |
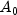 |
| |
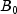 |
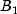 |
 |
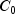 |
88.76 |
91.41 |
97.85 |
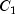 |
87.45 |
98.27 |
95.85 |
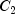 |
86.01 |
104.20 |
90.09 |
| |
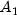 |
| |
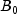 |
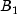 |
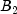 |
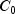 |
94.83 |
100.49 |
99.75 |
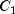 |
84.57 |
97.20 |
112.30 |
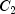 |
81.06 |
120.80 |
108.77 |
| |
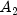 |
| |
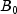 |
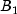 |
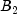 |
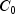 |
99.90 |
100.23 |
104.50 |
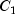 |
92.98 |
107.77 |
110.94 |
 |
94.72 |
118.39 |
102.87 |
import java.text.*;
import com.imsl.stat.*;
import com.imsl.math.PrintMatrix;
import com.imsl.math.PrintMatrixFormat;
public class ANOVAFactorialEx3 {
public static void main(String args[]) {
int nSubscripts = 3, i;
int[] nLevels = {3, 3, 3};
double[] y = {88.76, 87.45, 86.01, 91.41, 98.27, 104.2, 97.85, 95.85,
90.09, 94.83, 84.57, 81.06, 100.49, 97.2, 120.8, 99.75, 112.3, 108.77,
99.9, 92.98, 94.72, 100.23, 107.77, 118.39, 104.51, 110.94, 102.87};
String[] labels = {
"degrees of freedom for the model ",
"degrees of freedom for error ",
"total (corrected) degrees of freedom ",
"sum of squares for the model ",
"sum of squares for error ",
"total (corrected) sum of squares ",
"model mean square ",
"error mean square ",
"F-statistic ",
"p-value ",
"R-squared (in percent) ",
"Adjusted R-squared (in percent) ",
"est. standard deviation of the model error ",
"overall mean of y ",
"coefficient of variation (in percent) "
};
String[] rlabels = {"A", "B", "C", "A*B", "A*C", "B*C"};
NumberFormat nf = NumberFormat.getInstance();
ANOVAFactorial af = new ANOVAFactorial(nSubscripts, nLevels, y);
af.setErrorIncludeType(ANOVAFactorial.POOL_INTERACTIONS);
nf.setMinimumFractionDigits(6);
System.out.println("P-value = " + nf.format(af.compute()));
nf.setMaximumFractionDigits(4);
System.out.println("\n * * * Analysis of Variance * * *");
double[] anova = af.getANOVATable();
for (i = 0; i < anova.length; i++) {
System.out.println(labels[i] + " " + nf.format(anova[i]));
}
System.out.println("\n * * * Variation Due to the " +
"Model * * *");
System.out.println("Source\tDF\tSum of Squares\tMean Square" +
"\tProb. of Larger F");
double[][] te = af.getTestEffects();
for (i = 0; i < te.length; i++) {
StringBuffer sb = new StringBuffer(rlabels[i]);
int len = sb.length();
for(int j = 0; j < (8-len); j++) sb.append(' ');
sb.append(nf.format(te[i][0]));
len = sb.length();
for(int j = 0; j < (16-len); j++) sb.append(' ');
sb.append(nf.format(te[i][1]));
len = sb.length();
for(int j = 0; j < (32-len); j++) sb.append(' ');
sb.append(nf.format(te[i][2]));
len = sb.length();
for(int j = 0; j < (48-len); j++) sb.append(' ');
sb.append(nf.format(te[i][3]));
System.out.println(sb.toString());
}
}
}
Output
P-value = 0.008299
* * * Analysis of Variance * * *
degrees of freedom for the model 18.0000
degrees of freedom for error 8.0000
total (corrected) degrees of freedom 26.0000
sum of squares for the model 2,395.7290
sum of squares for error 185.7763
total (corrected) sum of squares 2,581.5052
model mean square 133.0961
error mean square 23.2220
F-statistic 5.7315
p-value 0.0083
R-squared (in percent) 92.8036
Adjusted R-squared (in percent) 76.6116
est. standard deviation of the model error 4.8189
overall mean of y 98.9619
coefficient of variation (in percent) 4.8695
* * * Variation Due to the Model * * *
Source DF Sum of Squares Mean Square Prob. of Larger F
A 2.0000 488.3675 10.5152 0.0058
B 2.0000 1,090.6564 23.4832 0.0004
C 2.0000 49.1485 1.0582 0.3911
A*B 4.0000 142.5853 1.5350 0.2804
A*C 4.0000 32.3474 0.3482 0.8383
B*C 4.0000 592.6238 6.3800 0.0131
Link to Java source.