Class ClusterHierarchical
- All Implemented Interfaces:
Serializable,Cloneable
Class ClusterHierarchical conducts a hierarchical cluster
analysis based upon a distance matrix, or, by appropriate use of the
transformation specified in the method setTransformType, based
upon a similarity matrix. Only the upper triangular part of the input matrix is used.
Hierarchical clustering in ClusterHierarchical proceeds as
follows:
Initially, each data point is considered to be a cluster, numbered 1 to n,
where n is the number of rows in the input matrix,
dist.
- If the input matrix contains similarities, the matrix is transformed
to a distance matrix using the transform type specified by the method
setTransformType. Set k = 1. - A search is made of the distance matrix to find the two closest clusters. These clusters are merged to form a new cluster, numbered n + k. The cluster numbers of the two clusters joined at this stage are saved as Right Sons and Left Sons, and the distance measure between the two clusters is stored as Cluster Level.
- Based upon the method of clustering, updating of the distance
measure in the row and column of
distcorresponding to the new cluster is performed. - Set k = k + 1. If k is less than n, go to Step 2.
The five methods differ primarily in how the distance matrix is updated
after two clusters have been joined. The argument method in
setMethod specifies how the distance of the cluster just
merged with each of the remaining clusters will be updated. Class
ClusterHierarchical allows five methods for computing the distances.
To understand these measures, suppose in the following discussion that clusters A
and B have just been joined to form cluster Z, and interest is in
computing the distance of Z with another cluster called C.
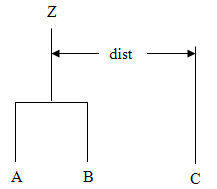
method |
Description |
LINKAGE_SINGLE | Single linkage (minimum distance). The distance from Z to C is the minimum of the distances (A to C, B to C). |
LINKAGE_COMPLETE | Complete linkage (maximum distance). The distance from Z to C is the maximum of the distances (A to C, B to C). |
LINKAGE_AVG_WITHIN_CLUSTERS | Average-distance-within-clusters method. The distance from Z to C is the average distance of all objects that would be within the cluster formed by merging clusters Z and C. This average may be computed according to formulas given by Anderberg (1973, page 139). |
LINKAGE_AVG_BETWEEN_CLUSTERS | Average-distance-between-clusters method. The distance from Z to C is the average distance of objects within cluster Z to objects within cluster C. This average may be computed according to methods given by Anderberg (1973, page 140). |
LINKAGE_WARDS | Ward's method: Clusters are formed so as to minimize the increase in the within-cluster sums of squares. The distance between two clusters is the increase in these sums of squares if the two clusters were merged. A method for computing this distance from a squared Euclidean distance matrix is given by Anderberg (1973, pages 142-145). |
In general, single linkage will yield long thin clusters while complete linkage will yield clusters that are more spherical. Average linkage and Ward's linkage tend to yield clusters that are similar to those obtained with complete linkage.
Class ClusterHierarchical produces a unique
representation of the binary cluster tree via the following three
conventions; the fact that the tree is unique should aid in interpreting the
clusters. First, when two clusters are joined and each cluster contains two
or more data points, the cluster that was initially formed with the smallest
level becomes the left son. Second, when a cluster containing more than one
data point is joined with a cluster containing a single data point, the
cluster with the single data point becomes the right son. Finally, when two
clusters containing only one object are joined, the cluster with the
smallest cluster number becomes the right son.
Comments
- The clusters corresponding to the original data points are numbered
from 1 to
n, wherenis the number of rows indist. Then- 1 clusters formed by merging clusters are numberedn+ 1 ton+ (n- 1 ). - Raw correlations, if used as similarities, should be made positive
and transformed to a distance measure. One such transformation can
be performed by setting
transform=RECIPROCAL_ABS, in thesetTransformTypemethod. - The user may cluster either variables or observations with
ClusterHierarchicalsince a dissimilarity matrix, not the original data, is used. ClassDissimilaritiesmay be used to compute the matrixdistfor either the variables or observations.
- See Also:
-
Field Summary
FieldsModifier and TypeFieldDescriptionstatic final intIndicates the average distance between (average distance between objects in the two clusters) method.static final intIndicates the average distance within (average distance between objects within the merged cluster) method.static final intIndicates the complete linkage (maximum distance) method.static final intIndicates the single linkage (minimum distance) method.static final intIndicates the Ward's method.static final intIndicates transformation by multiplication by -1.0.static final intIndicates no transformation.static final intIndicates transformation by taking the reciprocal of the absolute value. -
Constructor Summary
ConstructorsConstructorDescriptionClusterHierarchical(double[][] dist) Constructor forClusterHierarchical.ClusterHierarchical(double[][] dist, int method, int transform) Deprecated. -
Method Summary
Modifier and TypeMethodDescriptionvoidcompute()Performs a hierarchical cluster analysis.final int[]Returns the left sons of each merged cluster.final double[]Returns the level at which the clusters are joined.final int[]getClusterMembership(int nClusters) Returns the cluster membership of each observation.final int[]Returns the right sons of each merged cluster.intReturns the clustering method used.final int[]getObsPerCluster(int nClusters) Returns the number of observations in each cluster.intReturns the type of transformation.voidsetMethod(int method) Sets the clustering method to be used.voidsetTransformType(int transform) Sets the type of transformation.
-
Field Details
-
LINKAGE_SINGLE
public static final int LINKAGE_SINGLEIndicates the single linkage (minimum distance) method.- See Also:
-
LINKAGE_COMPLETE
public static final int LINKAGE_COMPLETEIndicates the complete linkage (maximum distance) method.- See Also:
-
LINKAGE_AVG_WITHIN_CLUSTERS
public static final int LINKAGE_AVG_WITHIN_CLUSTERSIndicates the average distance within (average distance between objects within the merged cluster) method.- See Also:
-
LINKAGE_AVG_BETWEEN_CLUSTERS
public static final int LINKAGE_AVG_BETWEEN_CLUSTERSIndicates the average distance between (average distance between objects in the two clusters) method.- See Also:
-
LINKAGE_WARDS
public static final int LINKAGE_WARDSIndicates the Ward's method.- See Also:
-
NONE
public static final int NONEIndicates no transformation.- See Also:
-
MULTIPLICATION
public static final int MULTIPLICATIONIndicates transformation by multiplication by -1.0.- See Also:
-
RECIPROCAL_ABS
public static final int RECIPROCAL_ABSIndicates transformation by taking the reciprocal of the absolute value.- See Also:
-
-
Constructor Details
-
ClusterHierarchical
public ClusterHierarchical(double[][] dist, int method, int transform) Deprecated.UseClusterHierarchical(double[][])instead.Constructor forClusterHierarchical.- Parameters:
dist- Adoublesymmetric matrix containing the distance (or similarity) matrix. On input, only the upper triangular part needs to be present.ClusterHierarchicalsaves the upper triangular part ofdistin the lower triangle. On return, the upper triangular part ofdistis restored, and the matrix is made symmetric.method- Anintidentifying the clustering method to be used.methodDescription 0 Single linkage (minimum distance). 1 Complete linkage (maximum distance). 2 Average distance within (average distance between objects within the merged cluster). 3 Average distance between (average distance between objects in the two clusters). 4 Ward's method (minimize the within-cluster sums of squares). For Ward's method, the elements of distare assumed to be Euclidean distances.transform- Anintidentifying the type of transformation applied to the measures indist.transformDescription 0 No transformation is required. The elements of distare distances.1 Convert similarities to distances by multiplication by -1.0. 2 Convert similarities (usually correlations) to distances by taking the reciprocal of the absolute value. - Throws:
IllegalArgumentException- is thrown when the row lengths of input matrixaare not equal (i.e. the matrix edges are "jagged")
-
ClusterHierarchical
public ClusterHierarchical(double[][] dist) Constructor forClusterHierarchical.- Parameters:
dist- Adoublesymmetric matrix containing the distance (or similarity) matrix. Only the upper triangular part is used.
-
-
Method Details
-
setMethod
public void setMethod(int method) Sets the clustering method to be used.- Parameters:
method- Anintidentifying the clustering method to be used. By default,method=LINKAGE_SINGLE.methodDescription LINKAGE_SINGLESingle linkage (minimum distance). LINKAGE_COMPLETEComplete linkage (maximum distance). LINKAGE_AVG_WITHIN_CLUSTERSAverage distance within (average distance between objects within the merged cluster). LINKAGE_AVG_BETWEEN_CLUSTERSAverage distance between (average distance between objects in the two clusters). LINKAGE_WARDSWard's method (minimize the within-cluster sums of squares). For Ward's method, the elements of distare assumed to be Euclidean distances.
-
getMethod
public int getMethod()Returns the clustering method used. -
setTransformType
public void setTransformType(int transform) Sets the type of transformation.- Parameters:
transform- Anintidentifying the type of transformation applied to the measures indist. By default,transform=NONE.transformDescription NONENo transformation is required. The elements of distare distances.MULTIPLICATIONConvert similarities to distances by multiplication by -1.0. RECIPROCAL_ABSConvert similarities (usually correlations) to distances by taking the reciprocal of the absolute value.
-
getTransformType
public int getTransformType()Returns the type of transformation. -
compute
public void compute()Performs a hierarchical cluster analysis. -
getClusterLevel
public final double[] getClusterLevel()Returns the level at which the clusters are joined.- Returns:
- A
doublearray containing the level at which the clusters are joined. Element [k-1] contains the distance (or similarity) level at which clustern+ k was formed.
-
getClusterLeftSons
public final int[] getClusterLeftSons()Returns the left sons of each merged cluster.- Returns:
- An
intarray containing the left sons of each merged cluster.
-
getClusterRightSons
public final int[] getClusterRightSons()Returns the right sons of each merged cluster.- Returns:
- An
intarray containing the right sons of each merged cluster.
-
getClusterMembership
public final int[] getClusterMembership(int nClusters) Returns the cluster membership of each observation.- Parameters:
nClusters- Anintwhich specifies the desired number of clusters.- Returns:
- An
intarray containing the cluster membership of each observation.
-
getObsPerCluster
public final int[] getObsPerCluster(int nClusters) Returns the number of observations in each cluster.- Parameters:
nClusters- Anintwhich specifies the desired number of clusters.- Returns:
- An
intarray containing the number of observations in each cluster.
-
ClusterHierarchical(double[][])instead.