Analyzes categorical data using logistic, probit, Poisson, and other linear models.
For a list of all members of this type, see CategoricalGenLinModel Members.
System.Object
Imsl.Stat.CategoricalGenLinModel
Thread Safety
Public static (Shared in Visual Basic) members of this type are safe for multithreaded operations. Instance members are not guaranteed to be thread-safe.
Remarks
Reweighted least squares is used to compute (extended) maximum likelihood estimates in some generalized linear models involving categorized data. One of several models, including probit, logistic, Poisson, logarithmic, and negative binomial models, may be fit for input point or interval observations. (In the usual case, only point observations are observed.)
Let
CategoricalGenLinModel, observations with potentially infinite The models available in CategoricalGenLinModel are:
| Model Name | Parameterization | Response PDF |
|---|---|---|
| Model0 (Poisson) | ||
| Model1 (Negative Binomial) | ||
| Model2 (Logarithmic) | ||
| Model3 (Logistic) | ||
| Model4 (Probit) | ||
| Model5 (Log-log) |
Here ![]() denotes the cumulative normal distribution, N and S are known parameters specified for each observation via column OptionalDistributionParameterColumn of x, and w is an optional fixed parameter specified for each observation via column FixedParameterColumn of x. (By default N is taken to be 1 for model = 0, 3, 4 and 5 and S is taken to be 1 for model = 1. By default w is taken to be 0.) Since the log-log model (model = 5) probabilities are not symmetric with respect to 0.5, quantitatively, as well as qualitatively, different models result when the definitions of "success" and "failure" are interchanged in this distribution. In this model and all other models involving
denotes the cumulative normal distribution, N and S are known parameters specified for each observation via column OptionalDistributionParameterColumn of x, and w is an optional fixed parameter specified for each observation via column FixedParameterColumn of x. (By default N is taken to be 1 for model = 0, 3, 4 and 5 and S is taken to be 1 for model = 1. By default w is taken to be 0.) Since the log-log model (model = 5) probabilities are not symmetric with respect to 0.5, quantitatively, as well as qualitatively, different models result when the definitions of "success" and "failure" are interchanged in this distribution. In this model and all other models involving ![]() ,
, ![]() is taken to be the probability of a "success."
is taken to be the probability of a "success."
Note that each row vector in the data matrix can represent a single observation; or, through the use of column FrequencyColumn of the matrix x, each vector can represent several observations. Also note that classification variables and their products are easily incorporated into the models via the usual regression-type specifications.
Computational Details
For interval observations, the probability of the observation is computed by summing the probability distribution function over the range of values in the observation interval. For right-interval observations, ![]() is computed as a sum based upon the equality
is computed as a sum based upon the equality ![]() . Derivatives are similarly computed.
. Derivatives are similarly computed. CategoricalGenLinModel allows three types of interval observations. In full interval observations, both the lower and the upper endpoints of the interval must be specified. For right-interval observations, only the lower endpoint need be given while for left-interval observations, only the upper endpoint is given.
The computations proceed as follows:
- The input parameters are checked for consistency and validity.
- Estimates of the means of the "independent" or design variables are computed. The frequency of the observation in all but the binomial distribution model is taken from column
FrequencyColumnof the data matrix x. In binomial distribution models, the frequency is taken as the product of n =x[i,OptionalDistributionParameterColumn]andx[i,FrequencyColumn]. In all cases these values default to 1. Means are computed as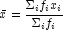
- If init = 0, initial estimates of the coefficients are obtained (based upon the observation intervals) as multiple regression estimates relating transformed observation probabilities to the observation design vector. For example, in the binomial distribution models,
 for point observations may be estimated as and, when model = 3, the linear relationship is given by
for point observations may be estimated as and, when model = 3, the linear relationship is given by![\hat{\theta}=x[i,LowerEndpointColumn]/x[i,
OptionalDistributionParameterColumn]](eqn/eqn_1839.png) while if
while if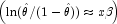
model= 4,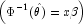
For bounded interval observations, the midpoint of the interval is used for
x[i,LowerEndpointColumn]. Right-interval observations are not used in obtaining initial estimates when the distribution has unbounded support (since the midpoint of the interval is not defined). When computing initial estimates, standard modifications are made to prevent illegal operations such as division by zero.Regression estimates are obtained at this point, as well as later, by use of linear regression.
- Newton-Raphson iteration for the maximum likelihood estimates is implemented via iteratively reweighted least squares. Let denote the log of the probability of the i-th observation for coefficients
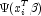
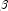 . In the least-squares model, the weight of the i-th observation is taken as the absolute value of the second derivative of with respect to
. In the least-squares model, the weight of the i-th observation is taken as the absolute value of the second derivative of with respect to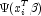 (times the frequency of the observation), and the dependent variable is taken as the first derivative
(times the frequency of the observation), and the dependent variable is taken as the first derivative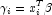
 with respect to
with respect to 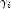 , divided by the square root of the weight times the frequency. The Newton step is given by where all derivatives are evaluated at the current estimate of
, divided by the square root of the weight times the frequency. The Newton step is given by where all derivatives are evaluated at the current estimate of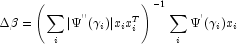
 , and
, and 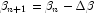 . This step is computed as the estimated regression coefficients in the least-squares model. Step halving is used when necessary to ensure a decrease in the criterion.
. This step is computed as the estimated regression coefficients in the least-squares model. Step halving is used when necessary to ensure a decrease in the criterion. - Convergence is assumed when the maximum relative change in any coefficient update from one iteration to the next is less than ConvergenceTolerance or when the relative change in the log-likelihood from one iteration to the next is less than
ConvergenceTolerance/100. Convergence is also assumed after MaxIterations or when step halving leads to a step size of less than .0001 with no increase in the log-likelihood. - For interval observations, the contribution to the log-likelihood is the log of the sum of the probabilities of each possible outcome in the interval. Because the distributions are discrete, the sum may involve many terms. The user should be aware that data with wide intervals can lead to expensive (in terms of computer time) computations.
If
InfiniteEstimateMethodis set to 0, then the methods of Clarkson and Jennrich (1991) are used to check for the existence of infinite estimates inAs an example of a situation in which infinite estimates can occur, suppose that observation j is right censored with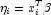
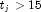 in a logistic model. If design matrix x is such that
in a logistic model. If design matrix x is such that 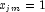 and
and 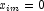 for all
for all 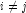 , then the optimal estimate of
, then the optimal estimate of 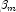 occurs at leading to an infinite estimate of both
occurs at leading to an infinite estimate of both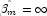
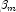 and
and  . In
. In CategoricalGenLinModel, such estimates may be "computed." In all models fit byCategoricalGenLinModel, infinite estimates can only occur when the optimal estimated probability associated with the left- or right-censored observation is 1. IfInfiniteEstimateMethodis set to 0, left- or right- censored observations that have estimated probability greater than 0.995 at some point during the iterations are excluded from the log-likelihood, and the iterations proceed with a log-likelihood based upon the remaining observations. This allows convergence of the algorithm when the maximum relative change in the estimated coefficients is small and also allows for the determination of observations with infiniteAt convergence, linear programming is used to ensure that the eliminated observations have infinite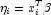
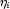 . If some (or all) of the removed observations should not have been removed (because their estimated
. If some (or all) of the removed observations should not have been removed (because their estimated 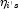 must be finite), then the iterations are restarted with a log-likelihood based upon the finite
must be finite), then the iterations are restarted with a log-likelihood based upon the finite 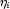 observations. See Clarkson and Jennrich (1991) for more details.
observations. See Clarkson and Jennrich (1991) for more details.
When
InfiniteEstimateMethodis set to 1, no observations are eliminated during the iterations. In this case, when infinite estimates occur, some (or all) of the coefficient estimates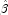 will become large, and it is likely that the Hessian will become (numerically) singular prior to convergence.
will become large, and it is likely that the Hessian will become (numerically) singular prior to convergence.When infinite estimates for the
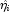 are detected, linear regression (see Chapter 2, Regression;) is used at the convergence of the algorithm to obtain unique estimates
are detected, linear regression (see Chapter 2, Regression;) is used at the convergence of the algorithm to obtain unique estimates 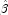 . This is accomplished by regressing the optimal
. This is accomplished by regressing the optimal 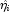 or the observations with finite
or the observations with finite  against
against 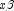 , yielding a unique
, yielding a unique 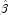 (by setting coefficients
(by setting coefficients 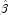 that are linearly related to previous coefficients in the model to zero). All of the final statistics relating to
that are linearly related to previous coefficients in the model to zero). All of the final statistics relating to 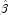 are based upon these estimates.
are based upon these estimates. Residuals are computed according to methods discussed by Pregibon (1981). Let
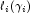 denote the log-likelihood of the i-th observation evaluated at
denote the log-likelihood of the i-th observation evaluated at 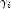 . Then, the standardized residual is computed as where
. Then, the standardized residual is computed as where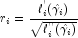
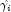 is the value of
is the value of 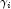 when evaluated at the optimal
when evaluated at the optimal 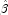 and the derivatives here (and only here) are with respect to
and the derivatives here (and only here) are with respect to  rather than with respect to
rather than with respect to 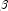 . The denominator of this expression is used as the "standard error of the residual" while the numerator is the "raw" residual.
. The denominator of this expression is used as the "standard error of the residual" while the numerator is the "raw" residual.
Following Cook and Weisberg (1982), we take the influence of the i-th observation to be
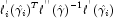
This quantity is a one-step approximation to the change in the estimates when the i-th observation is deleted. Here, the partial derivatives are with respect to
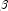 .
.
Programming Notes
- Classification variables are specified via ClassificationVariableColumn. Indicator or dummy variables are created for the classification variables.
- To enhance precision "centering" of covariates is performed if ModelIntercept is set to 1 and (number of observations) - (number of rows in x missing one or more values)
>1. In doing so, the sample means of the design variables are subtracted from each observation prior to its inclusion in the model. On convergence the intercept, its variance and its covariance with the remaining estimates are transformed to the uncentered estimate values. - Two methods for specifying a binomial distribution model are possible. In the first method,
x[i,FrequencyColumn]contains the frequency of the observation whilex[i,LowerEndpointColumn]is 0 or 1 depending upon whether the observation is a success or failure. In this case, N =x[i,OptionalDistributionParameterColumn]is always 1. The model is treated as repeated Bernoulli trials, and interval observations are not possible.
A second method for specifying binomial models is to use x[i,LowerEndpointColumn] to represent the number of successes in the x[i,OptionalDistributionParameterColumn] trials. In this case, x[i,FrequencyColumn] will usually be 1, but it may be greater than 1, in which case interval observations are possible.
Note that the Solve method must be called before using any property as a right operand, otherwise the value is null.
Requirements
Namespace: Imsl.Stat
Assembly: ImslCS (in ImslCS.dll)
See Also
CategoricalGenLinModel Members | Imsl.Stat Namespace | Example 1 | Example 2