lnormRegression¶
Fits a multiple linear regression model using either the Least Absolute Value (\(L_1\)), Least \(L_p\) norm (\(L_p\)), or Least Maximum Value (Minimax or \(L\infty\)) method of multiple linear regression.
Synopsis¶
lnormRegression (x, y)
Required Arguments¶
- float
x[[]](Input) - Array of size
nRows×nIndependentcontaining the independent (explanatory) variables(s). The i‑th column of x contains the i‑th independent variable. - float
y[](Input) - Array of size
nRowscontaining the dependent (response) variable.
Return Value¶
lnormRegression returns an array of length nIndependent + 1
containing a least absolute value solution for the regression coefficients.
The estimated intercept is the initial component of the array, where the
i‑th component contains the regression coefficients for the i‑th
dependent variable. If the optional argument noIntercept is used then
the (i-1)-st component contains the regression coefficients for the
i‑th dependent variable. lnormRegression returns the \(L_p\)
norm or least maximum value solution for the regression coefficients when
appropriately specified in the optional argument list.
Optional Arguments¶
methodLav (Input)
or
methodLlp, float (Input)
or
methodLmv, (Input)By default (or if
methodLavis specified) the function fits a multiple linear regression model using the least absolute values criterion.methodLlprequires the argument p, for \(p\geq 1\), and fits a multiple linear regression model using the \(L_p\) norm criterion.methodLmvfits a multiple linear regression model using the minimax criterion.
intercept (Input)
or
noIntercept, (Input)interceptis the default where the fitted value for observation i is\[\hat{\beta}_0 + \hat{\beta}_1 x_1 + \ldots + \hat{\beta}_k x_k\]where k =
nIndependent. IfnoInterceptis specified, the intercept term\[\left(\hat{\beta}_0\right)\]is omitted from the model and the return value from regression is an array of length
nIndependent.tolerance, float (Input)Tolerance used in determining linear dependence.
tolerance =100 ×machine(4)is the default.For more details see Chapter 15,:doc:/stat/utilities/index function machine.
weights, float[](Input)- Array of size
nRowscontaining the weights for the independent (explanatory) variable. frequencies, float[](Input)- Array of size
nRowscontaining the frequencies for the independent (explanatory) variable. maxIterations, int (Input)Maximum number of iterations allowed when using the multiple linear regression method.
maxIterationsis only applicable ifmethodLlpis specified.Default = 100
rank(Output)- Rank of the fitted model is returned in
rank. iterations(Output)- Number of iterations performed.
nRowsMissing(Output)- Number of rows of data containing NaN (not a number) for the dependent or independent variables. If a row of data contains NaN for any of these variables, that row is excluded from the computations.
sea(Output)- Sum of the absolute value of the errors.
seais only applicable ifmethodLavis also specified. maxResidual(Output)- Magnitude of the largest residual.
maxResidualis only applicable ifmethodLmvis specified. r(Output)- Upper triangular matrix of dimension (number of coefficients by number of
coefficients) containing the R matrix from a QR decomposition of the
matrix of regressors.
ris only applicable ifmethodLlpis specified. degreesOfFreedom(Output)- Sum of the frequencies minus
rank. In least squares fit (\(p=2\))degreesOfFreedomis called the degrees of freedom of error.degreesOfFreedomis only applicable ifmethodLlpis specified. residuals(Output)- An array of length
nRows(equal to the number of observations) containing the residuals.residualsis only applicable ifmethodLlpis specified. scale(Output)- Square of the scale constant used in an Lp analysis. An estimated
asymptotic variance-covariance matrix of the regression coefficients is
scale× \((R^T R)^{-1}\).scaleis only applicable ifmethodLlpis specified. residualsLpNorm(Output)- \(L_p\) norm of the residuals.
residualsLpNormis only applicable ifmethodLlpis specified. eps, float (Input)Convergence criterion. If the maximum relative difference in residuals from the k
-thto (k+1)-st iterations is less thaneps, convergence is declared.Default:
eps =100 ×machine(4).epsis only applicable ifmethodLlpis specified.
Description¶
Least Absolute Value Criterion¶
Function lnormRegression computes estimates of the regression
coefficients in a multiple linear regression model. For optional argument
lav (default), the criterion satisfied is the minimization of the sum of
the absolute values of the deviations of the observed response \(y_i\)
from the fitted response
for a set on n observations. Under this criterion, known as the \(L_1\) or LAV (least absolute value) criterion, the regression coefficient estimates minimize
The estimation problem can be posed as a linear programming problem. The special nature of the problem, however, allows for considerable gains in efficiency by the modification of the usual simplex algorithm for linear programming. These modifications are described in detail by Barrodale and Roberts (1973, 1974).
In many cases, the algorithm can be made faster by computing a least-squares
solution prior to the invocation of lav. This is particularly useful
when a least-squares solution has already been computed. The procedure is as
follows:
- Fit the model using least squares and compute the residuals from this fit.
- Fit the residuals from Step 1 on the regressor variables in the model
using
lav. - Add the two estimated regression coefficient vectors from Steps 1 and 2. The result is an \(L_1\) solution.
When multiple solutions exist for a given problem, option lav may yield
different estimates of the regression coefficients on different computers,
however, the sum of the absolute values of the residuals should be the same
(within rounding differences). The informational error indicating nonunique
solutions may result from rounding accumulation. Conversely, because of
rounding the error may fail to result even when the problem does have
multiple solutions.
\(L_p\) Norm Criterion¶
Optional argument llp computes estimates of the regression coefficients
in a multiple linear regression model \(y=X\beta+\varepsilon\) under the
criterion of minimizing the \(L_p\) norm of the deviations for
\(i=0,\ldots,n-1\) of the observed response \(y_i\) from the fitted
response
for a set on n observations and for \(p\geq 1\). For the case when
weights and frequencies are not supplied, the estimated regression
coefficient vector,
(output in coefficients[]) minimizes the \(L_p\) norm
The choice \(p=1\) yields the maximum likelihood estimate for β when the errors have a Laplace distribution. The choice \(p=2\) is best for errors that are normally distributed. Sposito (1989, pages 36−40) discusses other reasonable alternatives for p based on the sample kurtosis of the errors.
Weights are useful if the errors in the model have known unequal variances
In this case, the weights should be taken as
Frequencies are useful if there are repetitions of some observations in the
data set. If a single row of data corresponds to \(n_i\) observations,
set the frequency \(f_i=n_i\). In general, llp minimizes the
\(L_p\) norm
The asymptotic variance-covariance matrix of the estimated regression coefficients is given by
where R is from the QR decomposition of the matrix of regressors (output
in R-Matrix). An estimate of \(\lambda^2\) is output in scale.
In the discussion that follows, we will first present the algorithm with frequencies and weights all taken to be one. Later, we will present the modifications to handle frequencies and weights different from one.
Option call llp uses Newton’s method with a line search for p > 1.25
and, for p ≤ 1.25, uses a modification due to Ekblom (1973, 1987) in which
a series of perturbed problems are solved in order to guarantee convergence
and increase the convergence rate. The cutoff value of 1.25 as well as some
of the other implementation details given in the remaining discussion were
investigated by Sallas (1990) for their effect on CPU times.
In each case, for the first iteration a least-squares solution for the regression coefficients is computed using routine regression. If \(p=2\), the computations are finished. Otherwise, the residuals from the k-th iteration,
are used to compute the gradient and Hessian for the Newton step for the (k + 1)-st iteration for minimizing the p-th power of the \(L_p\) norm. (The exponent \(1/p\) in the \(L_p\) norm can be omitted during the iterations.)
For subsequent iterations, we first discuss the \(p>1.25\) case. For \(p>1.25\), the gradient and Hessian at the (k + 1)-st iteration depend upon
and
In the case 1.25 < p < 2 and
and the Hessian are undefined; and we follow the recommendation of Merle and Spath (1974). Specifically, we modify the definition of
to the following:
where \(\tau\) equals 100 × machine(4) times the square root of
the residual mean square from the least-squares fit. (See routine
machine which is documented in the section “Machine-Dependent Constants”
in Reference Material.)
Let \(\nu^{(k+1)}\) be a diagonal matrix with diagonal entries
and let \(z^{(k+1)}\) be a vector with elements
In order to compute the step on the (k + 1)-st iteration, the R from the QR decomposition of
is computed using fast Givens transformations. Let
denote the upper triangular matrix from the QR decomposition. The linear system
is solved for
where \(R^{(k+1)}\) is from the QR decomposition of \(\left[ v^{(k+1)}\right]^{1/2} X\). The step taken on the (k + 1)-st iteration is
The first attempted step on the (k + 1)-st iteration is with \(\alpha^{(k+1)}\). If all of the
are nonzero, this is exactly the Newton step. See Kennedy and Gentle (1980, pages 528−529) for further discussion.
If the first attempted step does not lead to a decrease of at least one-tenth of the predicted decrease in the p-th power of the \(L_p\) norm of the residuals, a backtracking linesearch procedure is used. The backtracking procedure uses a one-dimensional quadratic model to estimate the backtrack constant p. The value of p is constrained to be no less that 0.1.
An approximate upper bound for p is 0.5. If after 10 successive backtrack
attempts, \(\alpha^{(k)}=p_1 p_2 \ldots p_{10}\) does not produce a step
with a sufficient decrease, then lnormRegression issues a message with
error code 5. For further details on the backtrack line-search procedure, see
Dennis and Schnabel (1983, pages 126−127).
Convergence is declared when the maximum relative change in the residuals
from one iteration to the next is less than or equal to eps. The
relative change
in the i‑th residual from iteration k to iteration \(k+1\) is computed as follows:
where s is the square root of the residual mean square from the least-squares fit on the first iteration.
For the case 1 ≤ p ≤ 1.25, we describe the modifications to the previous procedure that incorporate Ekblom’s (1973) results. A sequence of perturbed problems are solved with a successively smaller perturbation constant c. On the first iteration, the least-squares problem is solved. This corresponds to an infinite c. For the second problem, c is taken equal to s, the square root of the residual mean square from the least-squares fit. Then, for the (j + 1)-st problem, the value of c is computed from the previous value of c according to
Each problem is stated as
For each problem, the gradient and Hessian on the (k + 1)-st iteration depend upon
and
where
The linear system \(\left[R^{(k+1)}\right]^T R^{(k+1)} d^{(k+1)}=X^T z^{(k+1)}\) is solved for \(d^{(k+1)}\) where \(R^{(k+1)}\) is from the QR decomposition of \(\left[V^{(k+1)}\right]^{1/2}X\). The step taken on the (k + 1)-st iteration is
where the first attempted step is with \(\alpha^{(k+1)}=1\). If necessary, the backtracking line-search procedure discussed earlier is used.
Convergence for each problem is relaxed somewhat by using a convergence
epsilon equal to max(eps, \(10^{-j}\)) where \(j=1,2,3,\ldots\)
indexes the problems (\(j=0\) corresponds to the least-squares problem).
After the convergence of a problem for a particular c, Ekblom’s (1987) extrapolation technique is used to compute the initial estimate of β for the new problem. Let \(R^{(k)}\),
and c be from the last iteration of the last problem. Let
and let t be the vector with elements \(t_i\). The initial estimate of β for the new problem with perturbation constant \(0.01c\) is
where \(\delta c=(0.01c-c)=-0.99c\), and where d is the solution of the linear system
Convergence of the sequence of problems is declared when the maximum
relative difference in residuals from the solution of successive problems is
less than eps.
The preceding discussion was limited to the case for which weights[i] =
1 and frequencies[i] = 1, i.e., the weights and frequencies are all
taken equal to one. The necessary modifications to the preceding algorithm
to handle weights and frequencies not all equal to one are as follows:
Replace
\[e_i^{(k)} \text{ by } \sqrt{w_i} e_i^{(k)}\]in the definitions of
\[z_i^{(k+1)}, v_i^{(k+1)}, \delta_i^{(k+1)}\]and \(t_i\).
Replace
These replacements have the same effect as multiplying the i‑th row of X and y by
and repeating the row \(f_i\) times except for the fact that the
residuals returned by lnormRegression are in terms of the original y
and X.
Finally, R and an estimate of \(\lambda^2\) are computed. Actually, R is recomputed because on output it corresponds to the R from the initial QR decomposition for least squares. The formula for the estimate of \(\lambda^2\) depends on p.
For \(p=1\), the estimator for \(\lambda^2\) is given by (McKean and Schrader 1987)
with
where \(z_{0.975}\) is the 97.5 percentile of the standard normal distribution, and where
are the ordered residuals where rank zero residuals are excluded. Note that
For \(p=2\), the estimator of \(\lambda^2\) is the customary least-squares estimator given by
For \(1<p<2\) and for \(p>2\), the estimator for \(\lambda^2\) is given by (Gonin and Money 1989)
with
Least Minimum Value Criterion (minimax)¶
Optional call lmv computes estimates of the regression coefficients in a
multiple linear regression model. The criterion satisfied is the
minimization of the maximum deviation of the observed response \(y_i\)
from the fitted response \(\hat{y}_i\) for a set on n observations.
Under this criterion, known as the minimax or LMV (least maximum value)
criterion, the regression coefficient estimates minimize
The estimation problem can be posed as a linear programming problem. A dual simplex algorithm is appropriate, however, the special nature of the problem allows for considerable gains in efficiency by modification of the dual simplex iterations so as to move more rapidly toward the optimal solution. The modifications are described in detail by Barrodale and Phillips (1975).
When multiple solutions exist for a given problem, lmv may yield
different estimates of the regression coefficients on different computers,
however, the largest residual in absolute value should have the same
absolute value (within rounding differences). The informational error
indicating nonunique solutions may result from rounding accumulation.
Conversely, because of rounding, the error may fail to result even when the
problem does have multiple solutions.
Examples¶
Example 1¶
A straight line fit to a data set is computed under the LAV criterion.
from __future__ import print_function
from numpy import *
from pyimsl.stat.lnormRegression import lnormRegression
xx = [1.0, 4.0, 2.0, 2.0, 3.0, 3.0, 4.0, 5.0]
yy = [1.0, 5.0, 0.0, 2.0, 1.5, 2.5, 2.0, 3.0]
sea = []
rank = []
iter = []
nrmiss = []
coefficients = lnormRegression(xx, yy,
sea=sea,
rank=rank,
iterations=iter,
nRowsMissing=nrmiss)
print("B = %6.2f\t%6.2f" % (coefficients[0], coefficients[1]))
print("Rank of Regressors Matrix = %3d" % (rank[0]))
print("Sum Absolute Value of Error = %8.4f" % (sea[0]))
print("Number of Iterations = %3d" % (iter[0]))
print("Number of Rows Missing = %3d" % (nrmiss[0]))
Output¶
B = 0.50 0.50
Rank of Regressors Matrix = 2
Sum Absolute Value of Error = 6.0000
Number of Iterations = 2
Number of Rows Missing = 0

Figure 2.2 — Least Squares and Least Absolute Value Fitted Lines
Example 2¶
Different straight line fits to a data set are computed under the criterion of minimizing the \(L_p\) norm by using p equal to 1, 1.5, 2.0 and 2.5.
from __future__ import print_function
from numpy import *
from pyimsl.stat.machine import machine
from pyimsl.stat.lnormRegression import lnormRegression
from pyimsl.stat.writeMatrix import writeMatrix
xx = [1.0, 4.0, 2.0, 2.0, 3.0, 3.0, 4.0, 5.0]
yy = [1.0, 5.0, 0.0, 2.0, 1.5, 2.5, 2.0, 3.0]
n_row = 2
n_col = 2
tolerance = 100 * machine(4)
convergence_eps = 0.001
p = 1.0
for i in range(0, 4):
eps = []
rank = []
iter = []
nrmiss = []
r_matrix = []
df_error = []
residuals = []
square_of_scale = []
lp_norm_residual = []
coefficients = lnormRegression(xx, yy,
methodLlp=p,
eps=convergence_eps,
rank=rank,
iterations=iter,
nRowsMissing=nrmiss,
r=r_matrix,
degreesOfFreedom=df_error,
residuals=residuals,
scale=square_of_scale,
residualsLpNorm=lp_norm_residual)
print("Coefficients = %6.2f\t%6.2f" % (coefficients[0], coefficients[1]))
print("Residuals = %6.2f\t%6.2f\t%6.2f\t%6.2f\t%6.2f\t%6.2f\t%6.2f\t%6.2f\n"
% (residuals[0], residuals[1], residuals[2], residuals[3], residuals[4],
residuals[5], residuals[6], residuals[7]))
print("P = %5.3f" % (p))
print("Lp norm of the residuals = %5.3f" % (lp_norm_residual[0]))
print("Rank of Regressors Matrix = %3d" % (rank[0]))
print("Degrees of Freedom Error = %5.3f" % (df_error[0]))
print("Number of Iterations = %3d" % (iter[0]))
print("Number of Missing Values = %3d" % (nrmiss[0]))
print("Square of Scale Constant = %5.3f" % (square_of_scale[0]))
writeMatrix("R Matrix", r_matrix)
print("---------------------------------------------------------")
p = p + 0.5
Output¶
Coefficients = 0.50 0.50
Residuals = -0.00 2.50 -1.50 0.50 -0.50 0.50 -0.50 -0.00
P = 1.000
Lp norm of the residuals = 6.002
Rank of Regressors Matrix = 2
Degrees of Freedom Error = 6.000
Number of Iterations = 8
Number of Missing Values = 0
Square of Scale Constant = 6.248
---------------------------------------------------------
Coefficients = 0.39 0.56
Residuals = 0.06 2.39 -1.50 0.50 -0.55 0.45 -0.61 -0.16
P = 1.500
Lp norm of the residuals = 3.712
Rank of Regressors Matrix = 2
Degrees of Freedom Error = 6.000
Number of Iterations = 6
Number of Missing Values = 0
Square of Scale Constant = 1.059
---------------------------------------------------------
Coefficients = -0.12 0.75
Residuals = 0.37 2.12 -1.38 0.62 -0.62 0.38 -0.88 -0.62
P = 2.000
Lp norm of the residuals = 2.937
Rank of Regressors Matrix = 2
Degrees of Freedom Error = 6.000
Number of Iterations = 1
Number of Missing Values = 0
Square of Scale Constant = 1.438
---------------------------------------------------------
Coefficients = -0.44 0.87
Residuals = 0.57 1.96 -1.30 0.70 -0.67 0.33 -1.04 -0.91
P = 2.500
Lp norm of the residuals = 2.540
Rank of Regressors Matrix = 2
Degrees of Freedom Error = 6.000
Number of Iterations = 4
Number of Missing Values = 0
Square of Scale Constant = 0.789
---------------------------------------------------------
R Matrix
1 2
1 2.828 8.485
2 0.000 3.464
R Matrix
1 2
1 2.828 8.485
2 0.000 3.464
R Matrix
1 2
1 2.828 8.485
2 0.000 3.464
R Matrix
1 2
1 2.828 8.485
2 0.000 3.464

Figure 2.3 — Various LP Fitted Lines
Example 3¶
A straight line fit to a data set is computed under the LMV criterion.
from __future__ import print_function
from numpy import *
from pyimsl.stat.lnormRegression import lnormRegression
xx = array([0.0, 1.0, 2.0, 3.0, 4.0, 4.0, 5.0])
yy = array([0.0, 2.5, 2.5, 4.5, 4.5, 6.0, 5.0])
max_residual = []
rank = []
itern = []
nrmiss = []
coefficients = lnormRegression(xx, yy,
methodLmv=True,
maxResidual=max_residual,
rank=rank,
iterations=itern,
nRowsMissing=nrmiss)
print("B = %6.2f\t%6.2f" % (coefficients[0], coefficients[1]))
print("Rank of Regressors Matrix = %3d" % (rank[0]))
print("Magnitude of Largest Residual = %8.4f" % (max_residual[0]))
print("Number of Iterations = %3d" % (itern[0]))
print("Number of Rows Missing = %3d" % (nrmiss[0]))
Output¶
B = 1.00 1.00
Rank of Regressors Matrix = 2
Magnitude of Largest Residual = 1.0000
Number of Iterations = 3
Number of Rows Missing = 0
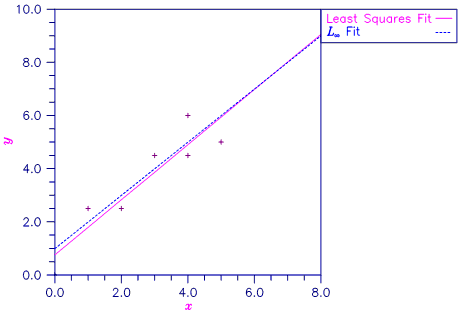
Figure 2.4 — Least Squares and Least Maximum Value