auto_parm
Estimates structural breaks in non-stationary univariate time series.
Synopsis
#include <imsls.h>
int *imsls_f_auto_parm (int nobs, float y[], int *npcs, ..., 0)
The type double function is imsls_d_auto_parm.
Required Arguments
int nobs (Input)
The number of observations in the time series (y).
The number of observations in the time series (y).
float y[] (Input)
An array of length nobs containing the time series.
An array of length nobs containing the time series.
int *npcs (Input/Output)
The number of requested/estimated pieces or segments of the time series. npcs is considered input only when IMSLS_AR_MODEL is provided.
The number of requested/estimated pieces or segments of the time series. npcs is considered input only when IMSLS_AR_MODEL is provided.
Return Value
A pointer to an array (arp) of length npcs × 2 containing the break points and AR orders for the derived model. If IMSLS_AR_MODEL is used, the return value is NULL.
Column Index | Description |
|---|---|
0 | Structural break points |
1 | AR order ( ) for each segment |
Synopsis with Optional Arguments
#include <imsls.h>
int *imsls_f_auto_parm (int nobs, float y[], int *npcs,
IMSLS_MAX_AR_ORDER, int max_ar_order,
IMSLS_METHOD, int method,
IMSLS_MODEL_SELECTION_CRITERION, int criterion,
IMSLS_MAXIMUM_LIKELIHOOD, int likelihood,
IMSLS_AR_MODEL, int arp[],
IMSLS_PRINT, int print,
IMSLS_RANDOM_SEED, int seed,
IMSLS_PROB_DISTRIBUTION, float pdistn[],
IMSLS_MIN_OBSERVATIONS, int mspan[],
IMSLS_GA_PARAMETERS, float gaparm[],
IMSLS_ISLAND, int island[],
IMSLS_MAX_MIGRATIONS, int maxmig,
IMSLS_STOP_ITERATIONS, int stopiters,
IMSLS_SELECTION_CRITERION_VALUE, float *value,
IMSLS_AR_FIT, float **arfit,
IMSLS_AR_FIT_USER, float arfit[],
IMSLS_AR_STATS, float **arstat,
IMSLS_AR_STATS_USER, float arstat[],
0)
Optional Arguments
IMSLS_MAX_AR_ORDER, int max_ar_order (Input)
Maximum order to consider for each AR model.
Maximum order to consider for each AR model.
Default: max_ar_order = 20.
IMSLS_METHOD, int method (Input)
Method of estimation.
Method of estimation.
method | Method Used |
|---|---|
0 | Yule –Walker |
1 | Least Squares |
2 | Burg |
Default: method = 0.
IMSLS_MODEL_SELECTION_CRITERION, int criterion (Input)
Selection criterion.
Selection criterion.
criterion | Criterion Used |
|---|---|
0 | Minimum Description Length (MDL) |
1 | Akaike’s Information Criterion (AIC) |
Default: criterion = 0.
IMSLS_MAXIMUM_LIKELIHOOD, int likelihood (Input)
Likelihood computation method.
Likelihood computation method.
likelihood | Computation Method |
|---|---|
0 | Exact |
1 | Approximate |
Default: likelihood = 0.
IMSLS_AR_MODEL, int arp[] (Input)
A user specified array of length npcs × 2 containing the break points and AR orders. When this argument is used, only the AR parameters and quality of the fit are determined.
A user specified array of length npcs × 2 containing the break points and AR orders. When this argument is used, only the AR parameters and quality of the fit are determined.
Column Index | Description |
|---|---|
0 | Structural break points |
1 | AR order ( ) for each segment |
IMSLS_PRINT, int print (Input)
Printing option.
Printing option.
print | Action |
|---|---|
0 | No printing |
1 | Prints final results only |
2 | Prints intermediate and final results |
Default: print = 0.
IMSLS_RANDOM_SEED, int seed (Input)
Seed of the random number generator. For the same data and parameter settings, imsls_f_auto_parm will return the same results each time if a seed is set. If seed = 0, the system clock will be used to generate a seed. The result will be nondeterministic.
Seed of the random number generator. For the same data and parameter settings, imsls_f_auto_parm will return the same results each time if a seed is set. If seed = 0, the system clock will be used to generate a seed. The result will be nondeterministic.
Default: seed = 0.
Note: The following input arguments are for setting up and running the embedded Genetic Algorithm. In most situations, the default values should be used for these arguments. Users may wish to change some or all for testing or research purposes. |
IMSLS_PROB_DISTRIBUTION, float pdistn[] (Input)
Array of length max_ar_order + 1 giving the probability distribution over the AR order variable p = 0,…, max_ar_order. i = 0,…, max_ar_order is used to randomly assign an AR order to breakpoint position for a given chromosome. pdistn[i] > = 0 and if Σpdistn is not equal to 1, the values will be normalized, i.e., pdistn[i] = pdistn[i]/ Σpdistn.
Array of length max_ar_order + 1 giving the probability distribution over the AR order variable p = 0,…, max_ar_order. i = 0,…, max_ar_order is used to randomly assign an AR order to breakpoint position for a given chromosome. pdistn[i] > = 0 and if Σpdistn is not equal to 1, the values will be normalized, i.e., pdistn[i] = pdistn[i]/ Σpdistn.
Default: pdistn[i] = 1/(max_ar_order + 1) for all i.
IMSLS_MIN_OBSERVATIONS, int mspan[] (Input)
Array of length max_ar_order + 1 containing minimum number of observations required for valid estimates of AR model with order p = 0, …, max_ar_order.
Array of length max_ar_order + 1 containing minimum number of observations required for valid estimates of AR model with order p = 0, …, max_ar_order.
Default: mspan [p] = 2 ×(number of parameters) + 2 = 2 × (p + 2) + 2.
IMSLS_GA_PARAMETERS, float gaparm[] (Input)
Array of length 4 containing parameters that control the behavior of the genetic algorithm. These values should be strictly greater than zero and less than one to avoid unexpected results.
Array of length 4 containing parameters that control the behavior of the genetic algorithm. These values should be strictly greater than zero and less than one to avoid unexpected results.
Index | Behavior |
|---|---|
0 | Probability used to set initial break points in a chromosome. Default: min (mspan) / nobs. |
1 | Probability of crossover used to decide between a crossover and a mutation. Default: 1 – min (mspan) / nobs. |
2 | In the mutation operation, probability an AR(p) model is enforced at the current position. Default: 0.4. |
3 | In the mutation operation, probability a break point is disallowed at the current position. Default: 0.3. |
Note: gaparm[2] and gaparm[3] must be valid probabilities and their sum must be between 0 and 1. 1 – gaparm[2] – gaparm[3] is the probability that the chromosome j inherits the parent's chromosome j. |
IMSLS_ISLAND, int island[] (Input)
Array of length 5 containing the migration policy parameters.
Array of length 5 containing the migration policy parameters.
Index | Policy |
|---|---|
0 | Number of islands. Default: 40. |
1 | Number of generations that pass before migration occurs. Note that the convergence of the algorithm is revised whether migrations take place or not (see argument island[4]). Default: 5. |
2 | Number of subjects that migrate at each migration event. Default: 2. |
3 | Population size (number of chromosomes) per island. Default: 40. |
4 | Migration flag. If 1, migration is performed. Default: 1. |
IMSLS_MAX_MIGRATIONS, int maxmig (Input)
Maximum number of times that migrations may take place before the function is stopped if convergence has not occurred.
Maximum number of times that migrations may take place before the function is stopped if convergence has not occurred.
Default: maxmig = 20.
IMSLS_STOP_ITERATIONS, int stopiters (Input)
Number of iterations. The function will declare convergence and stop the iterations if the criterion value (MDL/AIC) has not changed after stopiters consecutive migrations. Otherwise, the algorithm will declare non-convergence and stop after maxmig migrations have taken place. See also IMSLS_MAX_MIGRATIONS and island[1]. Note that logically, stopiters < maxmig.
Number of iterations. The function will declare convergence and stop the iterations if the criterion value (MDL/AIC) has not changed after stopiters consecutive migrations. Otherwise, the algorithm will declare non-convergence and stop after maxmig migrations have taken place. See also IMSLS_MAX_MIGRATIONS and island[1]. Note that logically, stopiters < maxmig.
Default: stopiters = 10.
IMSLS_SELECTION_CRITERION_VALUE, float *value (Output)
Final value of the selection criterion.
Final value of the selection criterion.
IMSLS_AR_FIT, float **arfit (Output)
Address of a pointer to an internally allocated array of length npcs × max_ar_order containing the AR coefficient estimates φ for each segment. arfit[i*max_ar_order+j] is the j‑th coefficient for segment i where i = 0, …, npcs - 1 and j = 0, …, arp[i×2 + 1].
Address of a pointer to an internally allocated array of length npcs × max_ar_order containing the AR coefficient estimates φ for each segment. arfit[i*max_ar_order+j] is the j‑th coefficient for segment i where i = 0, …, npcs - 1 and j = 0, …, arp[i×2 + 1].
Note that the intercept is not reported.
IMSLS_AR_FIT_USER, float arfit[] (Output)
Storage for array arfit is provided by the user. If npcs is output, the user should allocate (nobs ‑ 1) × max_ar_order to assure sufficient space. See IMSLS_AR_FIT.
Storage for array arfit is provided by the user. If npcs is output, the user should allocate (nobs ‑ 1) × max_ar_order to assure sufficient space. See IMSLS_AR_FIT.
IMSLS_AR_STATS, float **arstat (Output)
Address of a pointer to an internally allocated array of length npcs × 2.
Address of a pointer to an internally allocated array of length npcs × 2.
Column Index | Description |
|---|---|
0 | Likelihood values for each of the fitted AR models |
1 | Residual variances for each of the fitted AR models |
IMSLS_AR_STATS_USER, float arstat[] (Output)
Storage for array arstat is provided by the user. If npcs is output, the user should allocate (nobs ‑ 1) × 2 to assure sufficient space. See IMSLS_AR_STATS.
Storage for array arstat is provided by the user. If npcs is output, the user should allocate (nobs ‑ 1) × 2 to assure sufficient space. See IMSLS_AR_STATS.
Description
Function imsls_f_auto_parm estimates the structural breaks of a non-stationary time series using, with permission from the authors, the method developed by Davis et al (2006). imsls_f_auto_parm estimates a partition of the time index and models the time series in each segment as a separate auto-regressive (AR(p)) process. The function returns the estimated breakpoints, the estimated AR(p) models, and supporting statistics.
For the observed time series 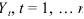 the problem is to find m, the number of breaks, their locations, , and , , the order of the AR process in which the j‑th segment is modeled. That is, for (for convenience, and ), where {Xt,j} is an AR process of order
the problem is to find m, the number of breaks, their locations, , and , , the order of the AR process in which the j‑th segment is modeled. That is, for (for convenience, and ), where {Xt,j} is an AR process of order
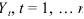 the problem is to find m, the number of breaks, their locations, , and , , the order of the AR process in which the j‑th segment is modeled. That is, for (for convenience, and ), where {Xt,j} is an AR process of order
the problem is to find m, the number of breaks, their locations, , and , , the order of the AR process in which the j‑th segment is modeled. That is, for (for convenience, and ), where {Xt,j} is an AR process of order and , the noise sequence, is independent and identically distributed with mean 0 and variance 1. Note that a series with m breaks will have m + 1 segments (m + 1 = npcs).
The vector completely specifies a piecewise AR model. To estimate this vector imsls_f_auto_parm optimizes, with respect to this vector, one of two selection criteria: the first is a Minimum Description Length (MDL) criterion, and the second is the Akaike's Information Criterion (AIC). The MDL is defined as
while the AIC criterion is given by
where, given a candidate value of the vector , is the likelihood of the fitted piecewise AR model evaluated at the parameter estimates,
The parameters of the j‑th AR segment are estimated by the choice of one of three estimation methods: Yule‑Walker, Burg, or Least Squares.
For simplicity, assume the mean of each series {Xt,j} is 0 and that the errors are Gaussian. Then, the piecewise AR model has Gaussian likelihood
where is the variance-covariance of the j‑th AR segment (of order ) and is the vector of observations of the j‑th segment, i.e., .
To find the minimizer of either MDL or AIC, imsls_f_auto_parm employs a Genetic Algorithm with islands, migration, cross-over and mutations. See Davis et.al. (2006) for further details.
Remarks
Function imsls_f_auto_parm approximates locally stationary time series by independent auto‑regressive processes. Experimental results suggest that imsls_f_auto_parm gives reasonable estimates of the structural breaks of a given time series, even if the segment series are not autoregressive. Also, based on experimental results, MDL gives better results than AIC as a selection criterion.
If seed is set out of range, an informational (error) message is issued indicating that the seed will be reset to 123457. Also if maxmig migrations are reached in the genetic algorithm before the selection criterion value converges an informational message is issued suggesting the increase of maxmig or the use of the double precision function.
Example
The examples below illustrate different scenarios using imsls_f_auto_parm. The example series used in each case is the airline demand data (Box, Jenkins and Reinsel, 1994), which gives monthly total demand for the period January 1949 through December 1960. Each scenario sets the optional argument, seed = 123457.
#include <imsls.h>
#include <stdio.h>
int main()
{
#define N 144
int n=N, npcs=0, iseed=123457, *arp=NULL, iper, iord, nlost;
int maxarorder, *arpnull=NULL, arp2[4]={0,2,59,1};
float x[N], *arfit=NULL, *arstat=NULL, sc, dx[N];
/* get data */
imsls_f_data_sets(4, IMSLS_RETURN_USER, x, 0);
/* Example 1: Use default values */
printf ("Example 1: Use defaults\n\n");
arp = imsls_f_auto_parm(n, x, &npcs,
IMSLS_PRINT, 1,
IMSLS_RANDOM_SEED, iseed,
IMSLS_SELECTION_CRITERION_VALUE, &sc,
IMSLS_AR_FIT, &arfit,
IMSLS_AR_STATS, &arstat,
0);
imsls_free(arp);
imsls_free(arfit);
imsls_free(arstat);
/* Example 2: differenced series set period for the difference.
iper is in years for this data set */
printf ("\n\nExample 2: differenced series\n\n");
iper = 1;
/* set the order for the difference. */
iord = 1;
/* get differenced series dx */
imsls_f_difference(n, x, 1, &iper,
IMSLS_ORDERS, &iord,
IMSLS_LOST, &nlost,
IMSLS_RETURN_USER, dx,
0);
arp = imsls_f_auto_parm(n-nlost, &dx[nlost], &npcs,
IMSLS_PRINT, 1,
IMSLS_RANDOM_SEED, iseed,
IMSLS_SELECTION_CRITERION_VALUE, &sc,
IMSLS_AR_FIT, &arfit,
IMSLS_AR_STATS, &arstat,
0);
imsls_free(arp);
imsls_free(arfit);
imsls_free(arstat);
/* Example 3: original series, lower order allowed
lower maximum AR order */
printf("\n\nExample 3: original series, lower order allowed\n\n");
maxarorder=5;
arp = imsls_f_auto_parm(n, x, &npcs,
IMSLS_MAX_AR_ORDER, maxarorder,
IMSLS_PRINT, 1,
IMSLS_RANDOM_SEED, iseed,
IMSLS_SELECTION_CRITERION_VALUE, &sc,
IMSLS_AR_FIT, &arfit,
IMSLS_AR_STATS, &arstat,
0);
imsls_free(arp);
imsls_free(arfit);
imsls_free(arstat);
/* Example 4: differenced series, lower order allowed */
printf("\n\nExample 4: differenced series, lower order allowed\n\n");
maxarorder=5;
arp = imsls_f_auto_parm(n-nlost, &dx[nlost], &npcs,
IMSLS_MAX_AR_ORDER, maxarorder,
IMSLS_PRINT, 1,
IMSLS_RANDOM_SEED, iseed,
IMSLS_SELECTION_CRITERION_VALUE, &sc,
IMSLS_AR_FIT, &arfit,
IMSLS_AR_STATS, &arstat,
0);
imsls_free(arp);
imsls_free(arfit);
imsls_free(arstat);
/* Example 5: original series, force fit the segments
Fit only at the break points */
printf("\n\nExample 5: original series, force fit the segments\n\n");
npcs=2;
arpnull = imsls_f_auto_parm(n, x, &npcs,
IMSLS_AR_MODEL, arp2,
IMSLS_PRINT, 1,
IMSLS_RANDOM_SEED, iseed,
IMSLS_SELECTION_CRITERION_VALUE, &sc,
IMSLS_AR_FIT, &arfit,
IMSLS_AR_STATS, &arstat,
0);
}
Output
Example 1: Use defaults
============== final results ===============
number of pieces: 2
selection criteria value: 684.243164
total time: 3.203000 conv: 1
==================== final model estimates =====================
break point order est. coeff. likelihood resid. var
arp[ 0] arp[ 1] arfit[ 0- 0] arstat[ 0] arstat[ 1]
0 1 0.77542
186.945 355.025
arp[ 2] arp[ 3] arfit[20-32] arstat[ 2] arstat[ 3]
43 13 1.03700
-0.07801
-0.03891
-0.03452
0.11961
-0.12851
0.01990
-0.04885
0.08089
-0.13117
0.22122
0.53862
-0.61515
486.666 691.486
Example 2: differenced series
============== final results ===============
number of pieces: 1
selection criteria value: 624.283508
total time: 3.031000 conv: 1
==================== final model estimates =====================
break point order est. coeff. likelihood resid. var
arp[ 0] arp[ 1] arfit[ 0-11] arstat[ 0] arstat[ 1]
0 12 -0.02842
-0.22436
-0.16846
-0.24267
-0.10573
-0.22429
-0.12126
-0.26446
-0.07087
-0.24327
-0.07136
0.57129
619.321 297.352
Example 3: original series, lower order allowed
============== final results ===============
number of pieces: 2
selection criteria value: 705.296631
total time: 2.312000 conv: 1
==================== final model estimates =====================
break point order est. coeff. likelihood resid. var
arp[ 0] arp[ 1] arfit[ 0- 0] arstat[ 0] arstat[ 1]
0 1 0.89533
270.393 333.563
arp[ 2] arp[ 3] arfit[ 5- 6] arstat[ 2] arstat[ 3]
62 2 1.19788
-0.35922
424.270 1632.335
Example 4: differenced series, lower order allowed
============== final results ===============
number of pieces: 2
selection criteria value: 698.359497
total time: 2.219000 conv: 1
==================== final model estimates =====================
break point order est. coeff. likelihood resid. var
arp[ 0] arp[ 1] arfit[ 0- 0] arstat[ 0] arstat[ 1]
0 0 -------
335.565 357.388
arp[ 2] arp[ 3] arfit[ 5- 5] arstat[ 2] arstat[ 3]
76 1 0.33310
352.175 1786.345
Example 5: original series, force fit the segments
============== final results ===============
number of pieces: 2
selection criteria value: 712.521
==================== final model estimates =====================
break point order est. coeff. likelihood resid. var
arp[ 0] arp[ 1] arfit[ 0- 1] arstat[ 0] arstat[ 1]
0 2 1.12156
-0.24876
258.192 313.889
arp[ 2] arp[ 3] arfit[20-20] arstat[ 2] arstat[ 3]
59 1 0.88605
443.696 1937.633
Warning Errors
IMSLS_MAX_MIGRATIONS_EXCEEDED | “maxmig” migrations or “stopiters” iterations were reached in the genetic algorithm before the selection criterion value converged. Try increasing “maxmig”, “stopiters” or using the double precision routine. |