Analyzes a balanced factorial design with fixed effects.
Synopsis
#include <imsls.h>
float imsls_f_anova_factorial (int n_subscripts, int n_levels, float y[], ..., 0)
The type double function is imsls_d_anova_factorial
Required Arguments
int
n_subscripts (Input)
Number of subscripts. Number of
factors in the model + 1 (for the error term).
int n_levels
(Input)
Array of length n_subscripts
containing the number of levels for each of the factors for the first n_subscripts − 1
elements. n_levels [n_subscripts − 1] is the
number of observations per cell.
float y[]
(Input)
Array of length n_levels [0]*n_levels [1]* … *n_levels [n_subscripts − 1]
containing the responses. Argument y must not contain NaN for any of its
elements, i.e., missing values are not allowed.
Return Value
The p-value for the overall F test.
Synopsis with Optional Arguments
#include <imsls.h>
float
imsls_f_anova_factorial (int
n_subscripts,
int
n_levels,
float
y[],
IMSLS_MODEL_ORDER, int
model_order,
IMSLS_PURE_ERROR, or
IMSLS_POOL_INTERACTIONS,
IMSLS_ANOVA_TABLE, float
**anova_table,
IMSLS_ANOVA_TABLE_USER, float
anova_table[],
IMSLS_TEST_EFFECTS, float
**test_effects,
IMSLS_TEST_EFFECTS_USER, float
test_effects[],
IMSLS_MEANS, float
**means,
IMSLS_MEANS_USER, float
means[],
0)
Optional Arguments
IMSLS_MODEL_ORDER, int
model_order (Input)
Number of factors to be included in
the highest-way interaction in the model. Argument model_order must be in
the interval [1, n_subscripts − 1]. For
example, a model_order of 1
indicates that a main effect model will be analyzed, and a model_order of 2
indicates that two-way interactions will be included in the model. Default:
model_order = n_subscripts − 1.
IMSLS_PURE_ERROR, or
IMSLS_POOL_INTERACTIONS
(Input)
IMSLS_PURE_ERROR,
the default option, indicates factor n_subscripts is error.
Its main effect and all its interaction effects are pooled into the error with
the other (model_order + 1)-way
and higher-way interactions. IMSLS_POOL_INTERACTIONS
indicates factor n_subscripts is not
error. Only (model_order + 1)-way
and higher-way interactions are included in the error.
IMSLS_ANOVA_TABLE, float
**anova_table (Output)
Address of a pointer to an
internally allocated array of size 15 containing the analysis of variance table.
The analysis of variance statistics are given as follows:
|
Element |
Analysis of Variance Statistics |
|
0 |
Degrees of freedom for the model. |
|
1 |
Degrees of freedom for error. |
|
2 |
Total (corrected) degrees of freedom. |
|
3 |
Sum of squares for the model. |
|
4 |
Sum of squares for error. |
|
5 |
Total (corrected) sum of squares. |
|
6 |
Model mean square. |
|
7 |
Error mean square. |
|
8 |
Overall F-statistic. |
|
9 |
p-value. |
|
10 |
R2 (in percent). |
|
11 |
Adjusted R2 (in percent). |
|
12 |
Estimate of the standard deviation. |
|
13 |
Overall mean of y. |
|
14 |
Coefficient of variation (in percent). |
Note that the p-value is returned as 0.0 when the value is so small that all significant digits have been lost.
IMSLS_ANOVA_TABLE_USER, float
anova_table[] (Output)
Storage for array anova_table is
provided by the user. See IMSLS_ANOVA_TABLE.
IMSLS_TEST_EFFECTS, float
**test_effects (Output)
Address of a pointer to an NEF × 4
internally allocated array containing a matrix containing statistics relating to
the sums of squares for the effects in the model. Here,
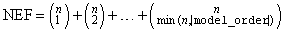
where n is given by n_subscripts if IMSLS_POOL_INTERACTIONS is specified; otherwise, n_subscripts − 1.
Suppose the factors are A, B, C, and error. With model_order = 3, rows 0 through NEF − 1 would correspond to A, B, C, AB, AC, BC, and ABC, respectively. The columns of test_effects are as follows:
|
Column |
Description |
|
0 |
Degrees of freedom. |
|
1 |
Sum of squares. |
|
2 |
F-statistic. |
|
3 |
p-value. |
Note that the p-value is returned as 0.0 when the value is so small that all significant digits have been lost.
IMSLS_TEST_EFFECTS_USER,
float test_effects[]
(Output)
Storage for array test_effects is provided by the user. See IMSLS_TEST_EFFECTS.
IMSLS_MEANS, float **means
(Output)
Address of a pointer to an internally allocated array of length
(n_levels [0] + 1) × (n_levels [1] + 1) × … ×
(n_levels[n − 1] + 1)
containing the subgroup means.
See argument IMSLS_TEST_EFFECTS for a definition of n. If the factors are A, B, C, and error, the ordering of the means is grand mean, A means, B means, C means, AB means, AC means, BC means, and ABC means.
IMSLS_MEANS_USER, float means[]
(Output)
Storage for array means is provided by
the user. See IMSLS_MEANS.
Description
Function imsls_f_anova_factorial performs an analysis for an n-way classification design with balanced data. For balanced data, there must be an equal number of responses in each cell of the n-way layout. The effects are assumed to be fixed effects. The model is an extension of the two-way model to include n factors. The interactions (two-way, three-way, up to n-way) can be included in the model, or some of the higher-way interactions can be pooled into error. The argument model_order specifies the number of factors to be included in the highest-way interaction. For example, if three-way and higher-way interactions are to be pooled into error, set model_order = 2. (By default, model_order = n_subscripts − 1 with the last subscript being the error subscript.) Argument IMSLS_PURE_ERROR indicates there are repeated responses within the n-way cell; IMSLS_POOL_INTERACTIONS_INTO_ERROR indicates otherwise.
Function imsls_f_anova_factorial requires the responses as input into a single vector y in lexicographical order, so that the response subscript associated with the first factor varies least rapidly, followed by the subscript associated with the second factor, and so forth. Hemmerle (1967, Chapter 5) discusses the computational method.
Examples
Example 1
A two-way analysis of variance is performed with balanced data discussed by Snedecor and Cochran (1967, Table 12.5.1, p. 347). The responses are the weight gains (in grams) of rats that were fed diets varying in the source (A) and level (B) of protein. The model is
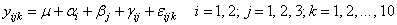
where
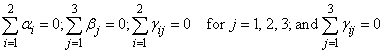
for i = 1, 2. The first responses in each cell in the two-way layout are given in the following table:
|
|
Protein Source (A) | ||
|
Protein Level (B) |
Beef |
Cereal |
Pork |
|
High |
73, 102, 118, 104, 81, 107, 100, 87, 117, 111 |
98, 74, 56, 111, 95, 88, 82, 77, 86, 92 |
94, 79, 96, 98, 102, 102, 108, 91, 120, 105 |
|
Low |
90, 76, 90, 64, 86, 51, 72, 90, 95, 78 |
107, 95, 97, 80, 98, 74, 74, 67, 89, 58 |
49, 82, 73, 86, 81, 97, 106, 70, 61, 82 |
#include <imsls.h>
int main ()
{
int n_subscripts= 3;
int n_levels[3] = {3,2,10};
float p_value;
float y[60] = {
73.0, 102.0, 118.0, 104.0, 81.0,
107.0, 100.0, 87.0, 117.0, 111.0,
90.0, 76.0, 90.0, 64.0, 86.0,
51.0, 72.0, 90.0, 95.0, 78.0,
98.0, 74.0, 56.0, 111.0, 95.0,
88.0, 82.0, 77.0, 86.0, 92.0,
107.0, 95.0, 97.0, 80.0, 98.0,
74.0, 74.0, 67.0, 89.0, 58.0,
94.0, 79.0, 96.0, 98.0, 102.0,
102.0, 108.0, 91.0, 120.0, 105.0,
49.0, 82.0, 73.0, 86.0, 81.0,
97.0, 106.0, 70.0, 61.0, 82.0};
p_value = imsls_f_anova_factorial(n_subscripts, n_levels, y, 0);
printf("P-value = %10.6f",p_value);
}
Output
P-value = 0.00229
Example 2
In this example, the same model and data is fit as in the initial example, but optional arguments are used for a more complete analysis.
#include <imsls.h>
int main ()
{
int n_subscripts= 3;
int n_levels[3] = {3,2,10};
float p_value;
float *test_effects, *means, *anova_table;
float y[60] = {
73.0, 102.0, 118.0, 104.0, 81.0,
107.0, 100.0, 87.0, 117.0, 111.0,
90.0, 76.0, 90.0, 64.0, 86.0,
51.0, 72.0, 90.0, 95.0, 78.0,
98.0, 74.0, 56.0, 111.0, 95.0,
88.0, 82.0, 77.0, 86.0, 92.0,
107.0, 95.0, 97.0, 80.0, 98.0,
74.0, 74.0, 67.0, 89.0, 58.0,
94.0, 79.0, 96.0, 98.0, 102.0,
102.0, 108.0, 91.0, 120.0, 105.0,
49.0, 82.0, 73.0, 86.0, 81.0,
97.0, 106.0, 70.0, 61.0, 82.0};
char *labels[] = {
"degrees of freedom for the model",
"degrees of freedom for error",
"total (corrected) degrees of freedom",
"sum of squares for the model",
"sum of squares for error",
"total (corrected) sum of squares",
"model mean square", "error mean square",
"F-statistic", "p-value",
"R-squared (in percent)","Adjusted R-squared (in percent)",
"est. standard deviation of the model error",
"overall mean of y",
"coefficient of variation (in percent)"};
char *test_row_labels[] = {"A", "B", "A*B"};
char *test_col_labels[] = {
"Source", "DF", "Sum of\nSquares",
"Mean\nSquare", "Prob. of\nLarger F"};
char *mean_row_labels[] = {
"grand mean",
"A1", "A2", "A3",
"B1", "B2",
"A1*B1", "A1*B2", "A2*B1", "A2*B2", "A3*B1", "A3*B2"};
/* Perform analysis */
p_value = imsls_f_anova_factorial(n_subscripts, n_levels, y,
IMSLS_ANOVA_TABLE, &anova_table,
IMSLS_TEST_EFFECTS, &test_effects,
IMSLS_MEANS, &means,
0);
printf("P-value = %10.6f",p_value);
/* Print results */
imsls_f_write_matrix("* * * Analysis of Variance * * *\n", 15, 1,
anova_table,
IMSLS_ROW_LABELS, labels,
IMSLS_WRITE_FORMAT, "%11.4f",
0);
imsls_f_write_matrix("* * * Variation Due to the Model * * *", 3, 4,
test_effects,
IMSLS_ROW_LABELS, test_row_labels,
IMSLS_COL_LABELS, test_col_labels,
IMSLS_WRITE_FORMAT, "%11.4f",
0);
imsls_f_write_matrix("* * * Subgroup Means * * *", 12, 1,
means,
IMSLS_ROW_LABELS, mean_row_labels,
IMSLS_WRITE_FORMAT, "%11.4f",
0);
}
Output
P-value = 0.002299
* * * Analysis of Variance * * *
degrees of freedom for the model 5.0000
degrees of freedom for error 54.0000
total (corrected) degrees of freedom 59.0000
sum of squares for the model 4612.9346
sum of squares for error 11585.9990
total (corrected) sum of squares 16198.9336
model mean square 922.5869
error mean square 214.5555
F-statistic 4.3000
p-value 0.0023
R-squared (in percent) 28.4768
Adjusted R-squared (in percent) 21.8543
est. standard deviation of the model error 14.6477
overall mean of y 87.8667
coefficient of variation (in percent) 16.6704
* * * Variation Due to the Model * * *
Source DF Sum of Mean Prob. Of
Squares Square Larger F
A 2.0000 266.5330 0.6211 0.5411
B 1.0000 3168.2678 14.7667 0.0003
A*B 2.0000 1178.1337 2.7455 0.0732
* * * Subgroup Means * * *
grand mean 87.8667
A1 89.6000
A2 84.9000
A3 89.1000
B1 95.1333
B2 80.6000
A1*B1 100.0000
A1*B2 79.2000
A2*B1 85.9000
A2*B2 83.9000
A3*B1 99.5000
A3*B2 78.7000
Example 3
This example performs a three-way analysis of variance using data discussed by Peter W.M. John (1971, pp. 91−92). The responses are weights (in grams) of roots of carrots grown with varying amounts of applied nitrogen (A), potassium (B), and phosphorus (C). Each cell of the three-way layout has one response. Note that the ABC interactions sum of squares, which is 186, is given incorrectly by Peter W.M. John (1971, Table 5.2.) The three-way layout is given in the following table:
|
|
A0 |
A1 |
A2 | |||||||||
|
|
B0 |
B1 |
B2 |
B0 |
B1 |
B2 |
B0 |
B1 |
B2 |
| ||
|
C0 |
88.76 |
91.41 |
97.85 |
94.83 |
100.49 |
99.75 |
99.90 |
100.23 |
104.51 |
| ||
|
C1 |
87.45 |
98.27 |
95.85 |
84.57 |
97.20 |
112.30 |
92.98 |
107.77 |
110.94 |
| ||
|
C2 |
86.01 |
104.20 |
90.09 |
81.06 |
120.80 |
108.77 |
94.72 |
118.39 |
102.87 |
| ||
#include <imsls.h>
int main ()
{
int n_subscripts= 3;
int n_levels[3] = {3,3,3};
float p_value;
float *test_effects, *anova_table;
float y[27] = {
88.76, 87.45, 86.01, 91.41, 98.27, 104.2, 97.85, 95.85,
90.09, 94.83, 84.57, 81.06, 100.49, 97.2, 120.8, 99.75,
112.3, 108.77, 99.9, 92.98, 94.72, 100.23, 107.77, 118.39,
104.51, 110.94, 102.87};
char *labels[] = {
"degrees of freedom for the model",
"degrees of freedom for error",
"total (corrected) degrees of freedom",
"sum of squares for the model",
"sum of squares for error",
"total (corrected) sum of squares",
"model mean square", "error mean square",
"F-statistic", "p-value",
"R-squared (in percent)","Adjusted R-squared (in percent)",
"est. standard deviation of the model error",
"overall mean of y",
"coefficient of variation (in percent)"};
char *test_row_labels[] = {"A", "B", "C", "A*B", "A*C", "B*C"};
char *test_col_labels[] = {
"Source", "DF", "Sum of\nSquares",
"Mean\nSquare", "Prob. of\nLarger F"};
/* Perform analysis */
p_value = imsls_f_anova_factorial(n_subscripts, n_levels, y,
IMSLS_ANOVA_TABLE, &anova_table,
IMSLS_TEST_EFFECTS, &test_effects,
IMSLS_POOL_INTERACTIONS,
0);
/* Print results */
printf("P-value = %10.6f",p_value);
imsls_f_write_matrix("* * * Analysis of Variance * * *\n", 15, 1,
anova_table,
IMSLS_ROW_LABELS, labels,
IMSLS_WRITE_FORMAT, "%11.4f",
0);
imsls_f_write_matrix("* * * Variation Due to the Model * * *", 6, 4,
test_effects,
IMSLS_ROW_LABELS, test_row_labels,
IMSLS_COL_LABELS, test_col_labels,
IMSLS_WRITE_FORMAT, "%11.4f",
0);
}
Output
P-value = 0.008299
* * * Analysis of Variance * * *
degrees of freedom for the model 18.0000
degrees of freedom for error 8.0000
total (corrected) degrees of freedom 26.0000
sum of squares for the model 2395.7290
sum of squares for error 185.7763
total (corrected) sum of squares 2581.5054
model mean square 133.0961
error mean square 23.2220
F-statistic 5.7315
p-value 0.0083
R-squared (in percent) 92.8036
Adjusted R-squared (in percent) 76.6116
est. standard deviation of the model error 4.8189
overall mean of y 98.9619
coefficient of variation (in percent) 4.8695
* * * Variation Due to the Model * * *
Source DF Sum of Mean Prob. Of
Squares Square Larger F
A 2.0000 488.3678 10.5152 0.0058
B 2.0000 1090.6559 23.4832 0.0004
C 2.0000 49.1484 1.0582 0.3911
A*B 4.0000 142.5856 1.5350 0.2804
A*C 4.0000 32.3474 0.3482 0.8383
B*C 4.0000 592.6240 6.3800 0.0131
