Compute a pooled variance-covariance from the observations.
Synopsis
#include <imsls.h>
float *imsls_f_pooled_covariances (int n_rows, int n_variables, float *x, int n_groups, ..., 0)
The type double function is imsls_d_pooled_covariances.
Required Argument
int n_rows
(Input)
Number of rows observations) in the input matrix x.
int
n_variables (Input)
Number of variables to be used in
computing the covariance matrix.
float *x
(Input)
A n_rows × n_variables + 1
matrix containing the data. The first n_variables columns
correspond to the variables, and the last column (column n_variables must
contain the group numbers).
int n_groups
(Input)
Number of groups in the data.
Return Value
Matrix of size n_variables by n_variables containing the matrix of covariances.
Synopsis with Optional Arguments
#include <imsls.h>
float
*imsls_f_pooled_covariances (int n_rows,
int
n_variables,
float x[], int n_groups,
IMSLS_X_COL_DIM,
int
x_col_dim,
IMSLS_X_INDICES, int igrp, int
ind[],
int
ifrq,
int
iwt,
IMSLS_IDO,
int ido,
IMSLS_ROWS_ADD, or
IMSLS_ROWS_DELETE,
IMSLS_GROUP_COUNTS,
int
**gcounts,
IMSLS_GROUP_COUNTS_USER, int
gcounts[],
IMSLS_SUM_WEIGHTS, float **sum_weights,
IMSLS_SUM_WEIGHTS_USER,
float
sum_weights[],
IMSLS_MEANS_USER, float means[],
IMSLS_U, float **u,
IMSLS_U_USER,
float
u[],
IMSLS_N_ROWS_MISSING,
int
*nrmiss,
IMSLS_RETURN_USER, float c[],
0)
Optional Arguments
IMSLS_X_COL_DIM, int x_col_dim
(Input)
Default: x_col_dim = n_variables + 1
IMSLS_X_INDICES, int igrp, int ind[], int ifrq, int iwt
(Input)
Each of the four arguments contains indices indicating column numbers
of x in which
particular types of data are stored. Columns are numbered 0 ... x_col_dim − 1.
Parameter igrp contains the index for the column of x in which the group numbers are stored.
Parameter ind contains the indices of the variables to be used in the analysis.
Parameters ifrq and iwt contain the column numbers of x in which the frequencies and weights, respectively, are stored. Set ifrq = −1 if there will be no column for frequencies. Set iwt = −1 if there will be no column for weights. Weights are rounded to the nearest integer. Negative weights are not allowed.
Defaults: igrp = n_variables, ind[ ] = 0, 1, …, n_variables − 1, ifrq = −1, and iwt = −1
IMSLS_IDO, int ido
(Input)
Processing option.
|
ido |
Action |
|
0 |
This is the only invocation; all the data are input at once. (Default) |
|
1 |
This is the first invocation with this data; additional calls will be made. Initialization and updating for the n_rows observations of x will be performed. |
|
2 |
This is an intermediate invocation; updating for the n_rows observations of x will be performed. |
|
3 |
All statistics are updated for the n_rows observations. The covariance matrix computed. |
Default: ido = 0
IMSLS_ROWS_ADD, or
IMSLS_ROWS_DELETE
By default (or if IMSLS_ROWS_ADD is
specified), the observations in x are added into the
analysis. If IMSLS_ROWS_DELETE is
specified, the observations are deleted from the analysis. If ido = 0,
these optional arguments are ignored (data is always added if there is only one
invocation).
IMSLS_GROUP_COUNTS, int **gcounts
(Output)
Address of a pointer to an integer array of length n_groups containing
the number of observations in each group. Array gcounts is updated
when ido is
equal to 0, 1, or 2.
IMSLS_GROUP_COUNTS_USER, int gcounts[]
(Output)
Storage for integer array gcounts is provided by
the user. See IMSLS_GROUP_COUNTS.
IMSLS_SUM_WEIGHTS, float
**sum_weights (Output)
Address of a pointer to an array of
length n_groups
containing the sum of the weights times the frequencies in the
groups.
IMSLS_SUM_WEIGHTS_USER, float
sum_weights[] (Output)
Storage for array sum_weights is
provided by the user. See IMSLS_SUM_WEIGHTS.
IMSLS_MEANS, float **means
(Output)
Address of a pointer to an array of size n_groups × n_variables. The
i-th row of means contains the
group i variable means.
IMSLS_MEANS_USER, float means[]
(Output)
Storage for array means is provided by
the user. See IMSLS_MEANS.
IMSLS_U, float **u
(Output)
Address of a pointer to an array of size n_variables × n_variables containing
the lower matrix U, the lower triangular for the pooled sample
cross-products matrix. U is computed from the pooled sample covariance
matrix, S (See the “Description” section below),
as S = UTU.
IMSLS_U_USER, float u[]
(Output)
Storage for array u is provided by the
user. See IMSLS_U.
IMSLS_N_ROWS_MISSING, int *nrmiss
(Output)
Number of rows of data encountered in calls to imsls_f_pooled_covariances
containing missing values (NaN) for any of the variables used.
IMSLS_RETURN_USER, float c[]
(Output)
If specified, c returns the
covariance matrix. Storage for array c is provided by the
user.
Description
Function imsls_f_pooled_covariances computes the pooled variance-covariance matrix from a matrix of observations. The within-groups means are also computed. Listwise deletion of missing values is assumed so that all observations used are complete; in any row of x, if any element of the observation is missing, the row is not used. Function imsls_f_pooled_covariances should be used whenever the user suspects that the data has been sampled from populations with different means but identical variance-covariance matrices. If these assumptions cannot be made, a different variance-covariance matrix should be estimated within each group.
By default, all observations are processed in one call to imsls_f_pooled_covariances. The computations are the same as if imsls_f_pooled_covariances were consecutively called with ido equal to 1, 2, and 3. For brevity, the following discusses the computations with ido > 0.
When ido = 1 variables are initialized, workspace is allocated and input variables are checked for errrors.
If n_rows ≠ 0 (for any value of ido), the group observation totals, Ti, for i = 1, …, g, where g is the number of groups, are updated for the n_rows observations in x. The group totals are computed as:
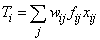
where wij is the observation weight, xij is the j-th observation in the i-th group, and fij is the observation frequency.
Modified Givens rotations are used in computed the Cholesky decomposition of the pooled sums of squares and crossproducts matrix. (Golub and Van Loan 1983).
The group means and the pooled sample covariance matrix S are computed from the intermediate results when ido = 3. These quantities are defined by
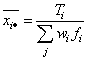
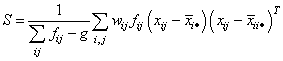
Examples
Example 1
The following example computes a pooled variance-covariance matrix. The last column of the data set is the group indicator.
#include <imsls.h>
int main() {
int nobs = 6;
int nvar = 2;
int n_groups = 2;
float *cov;
static float x[6][3] = {
2.2, 5.6, 1,
3.4, 2.3, 1,
1.2, 7.8, 1,
3.2, 2.1, 2,
4.1, 1.6, 2,
3.7, 2.2, 2};
cov = imsls_f_pooled_covariances(nobs, nvar, &x[0][0], n_groups, 0);
imsls_f_write_matrix("Pooled Covariance Matrix", nvar, nvar, cov, 0);
imsls_free(cov);
}
Output
Pooled Covariance Matrix
1 2
1 0.708 -1.575
2 -1.575 3.883
Example 2
The following example computes a pooled variance-covariance matrix for the Fisher iris data. To illustrate the use of the ido argument, multiple calls to imsls_f_pooled_covariances are made.
The first column of data is the group indicator, requiring either a permuation of the matrix or the use of the IMSLS_X_INDICES optional keyword. This example chooses the keyword method.
#include <imsls.h>
int main() {
int nobs = 150;
int nvar = 4;
int n_groups = 3;
int igrp = 0;
static int ind[4] = {1, 2, 3, 4};
int ifrq = -1;
int iwt = -1;
float *x, cov[16];
float *means;
int i;
/* Retrieve the Fisher iris data set */
x = imsls_f_data_sets(3, 0);
/* Initialize */
imsls_f_pooled_covariances(0, nvar, x, n_groups,
IMSLS_IDO, 1,
IMSLS_RETURN_USER, cov,
IMSLS_X_INDICES, igrp, ind, ifrq, iwt, 0);
/* Add 10 rows at a time */
for (i=0;i<15;i++) {
imsls_f_pooled_covariances(10, nvar, (x+i*50), n_groups,
IMSLS_IDO, 2,
IMSLS_RETURN_USER, cov,
IMSLS_X_INDICES, igrp, ind, ifrq, iwt, 0);
}
/* Calculate cov and free internal workspace */
imsls_f_pooled_covariances(0, nvar, x, n_groups,
IMSLS_IDO, 3,
IMSLS_RETURN_USER, cov,
IMSLS_X_INDICES, igrp, ind, ifrq, iwt,
IMSLS_MEANS, &means, 0);
imsls_f_write_matrix("Pooled Covariance Matrix", nvar, nvar, cov, 0);
imsls_f_write_matrix("Means", n_groups, nvar, means, 0);
imsls_free(means);
imsls_free(x);
}
Output
Pooled Covariance Matrix
1 2 3 4
1 0.2650 0.0927 0.1675 0.0384
2 0.0927 0.1154 0.0552 0.0327
3 0.1675 0.0552 0.1852 0.0427
4 0.0384 0.0327 0.0427 0.0419
Means
1 2 3 4
1 5.006 3.428 1.462 0.246
2 5.936 2.770 4.260 1.326
3 6.588 2.974 5.552 2.026
Warning Errors
IMSLS_OBSERVATION_IGNORED In call #, row # of the matrix “x” has group number = #. The group number must be between 1 and #, the number of groups. This observation will be ignored.
Fatal Errors
IMSLS_BAD_IDO_4 “ido” = #. Initial allocations must be performed by making a call to pooled_covariances with “ido” = 1.
IMSLS_BAD_IDO_5 “ido” = #. A new analysis may not begin until the previous analysis is terminated by a call to imsls_f_pooled_covariances with “ido” equal to 3.
