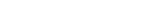SVGLM
Analyzes censored survival data using a generalized linear model.
Required Arguments
X — NOBS by NCOL matrix containing the data. (Input)
MODEL — Model option parameter. (Input)
MODEL specifies the distribution of the response variable and the relationship of the linear model to a distribution parameter.
MODEL specifies the distribution of the response variable and the relationship of the linear model to a distribution parameter.
MODEL | Distribution |
|---|---|
0 | Exponential |
1 | Linear hazard |
2 | Log‑normal |
3 | Normal |
4 | Log‑logistic |
5 | Logistic |
6 | Log least extreme value |
7 | Least extreme value |
8 | Log extreme value |
9 | Extreme value |
10 | Weibull |
For further discussion of the models and parameterizations used, see the “Description” section.
ILT — For interval‑censored and left‑censored observations, the column number in X that contains the upper endpoint of the failure interval. (Input)
See argument ICEN. If ILT = 0, left‑censored and interval‑censored observations cannot be input.
See argument ICEN. If ILT = 0, left‑censored and interval‑censored observations cannot be input.
IRT — For interval‑censored and right‑censored observations, the column number in X that contains the lower endpoint of the failure interval. (Input)
For exact‑failure observations, X(i, IRT) contains the exact‑failure time. IRT must not be zero. See argument ICEN.
For exact‑failure observations, X(i, IRT) contains the exact‑failure time. IRT must not be zero. See argument ICEN.
MAXCL — An upper bound on the sum of the number of distinct values taken by the classification variables. (Input)
NCOEF — Number of estimated coefficients in the model. (Output, if INIT = 0; Input, if INIT = 1)
COEF — NCOEF by 4 matrix containing parameter estimates and associated statistics. (Output, if INIT = 0; Input/Output, if INIT = 1; Input, if MAXIT = 0)
Col. | Statistic |
|---|---|
1 | Coefficient estimate. |
2 | Estimated standard deviation of the estimated coefficient. |
3 | Asymptotic normal score for testing that the coefficient is zero. |
4 | p‑value associated with the normal score in column 3. |
When COEF is input, only column 1 is referenced as input data, and columns 2 to 4 need not be set. When present in the model, the initial coefficient in COEF estimates a “nuisance” parameter, and the remaining coefficients estimate parameters associated with the “linear” model, beginning with the intercept, if present. Nuisance parameters are as follows:
Model | Nuisance Parameter |
|---|---|
1 | Coefficient of the quadratic term in time, θ |
2 – 9 | Scale parameter, σ |
10 | Shape parameter, θ |
ALGL — Maximized log‑likelihood. (Output)
COV — NCOEF by NCOEF matrix containing the estimated asymptotic covariance matrix of the coefficients. (Output)
COV is computed as the inverse of the matrix of second partial derivatives of negative one times the log‑likelihood. When MAXIT = 0, COV is computed at the initial estimates.
COV is computed as the inverse of the matrix of second partial derivatives of negative one times the log‑likelihood. When MAXIT = 0, COV is computed at the initial estimates.
XMEAN — Vector of length NCOEF containing the means of the design variables. (Output)
CASE — NOBS by 5 vector containing the case analysis. (Output)
Col. | Statistics |
|---|---|
1 | Estimated predicted value |
2 | Estimated influence or leverage |
3 | Residual estimate |
4 | Estimated cumulative hazard |
5 | For non‑censored observations, the estimated density at the observation failure time and covariate values. For censored observations, the corresponding estimated probability. |
If MAXIT = 0, CASE is a NOBS by 1 vector containing the estimated probability (for censored observations) or the estimated density (for non censored observations).
GR — Vector of length NCOEF containing the last parameter updates, excluding step halvings. (Output)
GR is computed as the inverse of the matrix of second partial derivatives times the vector of first partial derivatives of the log‑likelihood. When MAXIT = 0, the derivatives are computed at the initial estimates.
GR is computed as the inverse of the matrix of second partial derivatives times the vector of first partial derivatives of the log‑likelihood. When MAXIT = 0, the derivatives are computed at the initial estimates.
IADDS — Vector of length NOBS indicating which observations have and have not been included in the model. (Output, if MAXIT > 0; Input/Output, if MAXIT = 0)
Value | Status of Observation |
|---|---|
0 | Observation i has been included in the model. |
1 | Observation i has not been included in the model due to missing values in the X matrix. |
2 | Observation i has not been included in the model because of infinite estimates in extended maximum likelihood estimation. If MAXIT = 0, then the IADDS array must be initialized prior to calling SVGLM. |
Optional Arguments
NOBS — Number of observations. (Input)
Default: NOBS = size (X,1).
Default: NOBS = size (X,1).
NCOL — Number of columns in X. (Input)
Default: NCOL = size (X,2).
Default: NCOL = size (X,2).
LDX — Leading dimension of X exactly as specified in the dimension statement in the calling program. (Input)
Default: LDX = size (X,1).
Default: LDX = size (X,1).
IFRQ — Column number in X containing the frequency of response for each observation. (Input)
If IFRQ = 0, a response frequency of 1 for each observation is assumed.
Default: IFRQ = 0.
If IFRQ = 0, a response frequency of 1 for each observation is assumed.
Default: IFRQ = 0.
IFIX — Column number in X containing a constant to be added to the linear response. (Input)
Default: IFIX = 0.
The estimated linear response is taken to be where wi is the observation constant, zi is the observation design vector, is the vector of estimated parameters output in the first column of COEF, and i indexes the observations. The “fixed” constant allows one to test hypotheses about parameters via the log‑likelihoods. If IFIX = 0, the fixed parameter is assumed to be 0.
Default: IFIX = 0.
The estimated linear response is taken to be where wi is the observation constant, zi is the observation design vector, is the vector of estimated parameters output in the first column of COEF, and i indexes the observations. The “fixed” constant allows one to test hypotheses about parameters via the log‑likelihoods. If IFIX = 0, the fixed parameter is assumed to be 0.
ICEN — Column number in X containing the censoring code for each observation. (Input)
Default: ICEN = 0.
If ICEN = 0, a censoring code of 0 is assumed. Valid censoring codes are:
Default: ICEN = 0.
If ICEN = 0, a censoring code of 0 is assumed. Valid censoring codes are:
X(i, ICEN) | Censoring |
|---|---|
0 | Exact failure at X(i, IRT). |
1 | Right censored. The response is greater than X(i, IRT). |
2 | Left censored. The response is less than or equal to X(i, ILT). |
3 | Interval censored. The response is greater than X(i, IRT), but less than or equal to X(i, ILT). |
INFIN — Method to be used for handling infinite estimates. (Input)
Default: INFIN = 0.
Default: INFIN = 0.
INFIN | Method |
|---|---|
0 | Remove a right or left‑censored observation from the loglikelihood whenever the probability of the observation exceeds 0.995. At convergence, use linear programming to check that all removed observations actually have infinite linear response |
. | |
Set IADDS(i) for observation i to 2 if the linear response is infinite. If not all removed observations have infinite linear response | |
. | |
1 | Iterate without checking for infinite estimates. |
See the “Description” section for more discussion.
MAXIT — Maximum number of iterations. (Input)
MAXIT = 30 will usually be sufficient. Use MAXIT = 0 to compute the Hessian and score vector at the initial estimates.
Default: MAXIT = 30.
MAXIT = 30 will usually be sufficient. Use MAXIT = 0 to compute the Hessian and score vector at the initial estimates.
Default: MAXIT = 30.
EPS — Convergence criterion. (Input)
Convergence is assumed when the maximum relative change in any coefficient estimate is less than EPS from one iteration to the next, or when the relative change in the log‑likelihood, ALGL, from one iteration to the next is less than EPS/100. If EPS is negative, EPS = 0.001 is assumed.
Default: EPS = 0.001.
Convergence is assumed when the maximum relative change in any coefficient estimate is less than EPS from one iteration to the next, or when the relative change in the log‑likelihood, ALGL, from one iteration to the next is less than EPS/100. If EPS is negative, EPS = 0.001 is assumed.
Default: EPS = 0.001.
INTCEP — Intercept option. (Input)
Default: INTCEP = 1.
Default: INTCEP = 1.
INTCEP | Action |
|---|---|
0 | No intercept is in the model (unless otherwise provided for by the user). |
1 | An intercept is automatically included in the model.. |
NCLVAR — Number of classification variables. (Input)
Dummy or indicator variables are generated for classification variables using the IDUMMY = 2 option of routine GRGLM (see Chapter 2, “Regression”). See Comment 3.
Default: NCLVAR = 0.
Dummy or indicator variables are generated for classification variables using the IDUMMY = 2 option of routine GRGLM (see Chapter 2, “Regression”). See Comment 3.
Default: NCLVAR = 0.
INDCL — Index vector of length NCLVAR containing the column numbers of X that are classification variables. (Input, if NCLVAR is positive, not used otherwise)
If NCLVAR is 0, INDCL is not referenced and can be dimensioned of length 1 in the calling program.
If NCLVAR is 0, INDCL is not referenced and can be dimensioned of length 1 in the calling program.
NEF — Number of effects in the model. (Input)
In addition to effects involving classification variables, simple covariates and the product of simple covariates are also considered effects.
Default: NEF = 0.
In addition to effects involving classification variables, simple covariates and the product of simple covariates are also considered effects.
Default: NEF = 0.
NVEF — Vector of length NEF containing the number of variables associated with each effect in the model. (Input, if NEF is positive; not used otherwise)
If NEF is zero, NVEF is not used and can be dimensioned of length 1 in the calling program.
If NEF is zero, NVEF is not used and can be dimensioned of length 1 in the calling program.
INDEF — Index vector of length NVEF(1) + NVEF(2) + … + NVEF(NEF) containing the column numbers in X associated with each effect. (Input, if NEF is positive; not used otherwise)
The first NVEF(1) elements of INDEF give the column numbers in X of the variables in the first effect. The next NVEF(2) elements of INDEF give the column numbers for the second effect, etc. If NEF is zero, INDEF is not used and can be dimensioned of length one in the calling program.
The first NVEF(1) elements of INDEF give the column numbers in X of the variables in the first effect. The next NVEF(2) elements of INDEF give the column numbers for the second effect, etc. If NEF is zero, INDEF is not used and can be dimensioned of length one in the calling program.
INIT — Initialization option. (Input)
Default: INIT = 0.
Default: INIT = 0.
INIT | Action |
|---|---|
0 | Unweighted linear regression is used to obtain initial estimates. |
1 | The NCOEF elements in the first column of COEF contain initial estimates of the parameters on input to SVGLM (requiring that the user know NCOEF prior to calling SVGLM). |
IPRINT — Printing option. (Input)
Default: IPRINT = 0.
Default: IPRINT = 0.
IPRINT | Action |
|---|---|
0 | No printing is performed.. |
1 | Printing is performed, but observational statistics are not printed.. |
2 | All output statistics are printed. |
NCLVAL — Vector of length NCLVAR containing the number of values taken by each classification variable. (Output, if NCLVAR is positive; not used otherwise)
NCLVAL(i) is the number of distinct values for the i‑th classification variable. If NCLVAR is zero, NCLVAL is not used and can be dimensioned of length 1 in the calling program.
NCLVAL(i) is the number of distinct values for the i‑th classification variable. If NCLVAR is zero, NCLVAL is not used and can be dimensioned of length 1 in the calling program.
CLVAL — Vector of length NCLVAL(1) + NCLVAL(2) + … + NCLVAL(NCLVAR) containing the distinct values of the classification variables in ascending order. (Output, if NCLVAR is positive; not used otherwise)
The first NCLVAL(1) elements contain the values for the first classification variables, the next NCLVAL(2) elements contain the values for the second classification variable, etc. If NCLVAR is zero, then CLVAL is not referenced and can be dimensioned of length 1 in the calling program.
The first NCLVAL(1) elements contain the values for the first classification variables, the next NCLVAL(2) elements contain the values for the second classification variable, etc. If NCLVAR is zero, then CLVAL is not referenced and can be dimensioned of length 1 in the calling program.
LDCOEF — Leading dimension of COEF exactly as specified in the dimension statement in the calling program. (Input)
Default: LDCOEF = size (COEF,1).
Default: LDCOEF = size (COEF,1).
LDCOV — Leading dimension of COV exactly as specified in the dimension statement in the calling program. (Input)
Default: LDCOV = size (COV,1).
Default: LDCOV = size (COV,1).
LDCASE — Leading dimension of CASE exactly as specified in the dimension statement in the calling program. (Input)
Default: LDCASE = size (CASE,1).
Default: LDCASE = size (CASE,1).
NRMISS — Number of rows of data in X that contain missing values in one or more columns ILT, IRT, IFRQ, ICOEF, ICEN, INDCL or INDEF of X. (Output)
FORTRAN 90 Interface
Generic: CALL SVGLM (X, MODEL, ILT, IRT, MAXCL, NCOEF, COEF, ALGL, COV, XMEAN, CASE, GR, IADDS [, …])
Specific: The specific interface names are S_SVGLM and D_SVGLM.
FORTRAN 77 Interface
Single: CALL SVGLM (NOBS, NCOL, X, LDX, MODEL, ILT, IRT, IFRQ, IFIX, ICEN, INFIN, MAXIT, EPS, INTCEP, NCLVAR, INDCL, NEF, NVEF, INDEF, INIT, IPRINT, MAXCL, NCLVAL, CLVAL, NCOEF, COEF, LDCOEF, ALGL, COV, LDCOV, XMEAN, CASE, LDCASE, GR, IADDS, NRMISS)
Double: The double precision name is DSVGLM.
Description
Routine SVGLM computes maximum likelihood estimates of parameters and associated statistics in generalized linear models commonly found in survival (reliability) analysis. Although the terminology used will be from the survival area, the methods discussed have application in many areas of data analysis, including reliability analysis and event history analysis. Indeed, these methods may be used anywhere a random variable from one of the discussed distributions is parameterized via one of the models available in SVGLM. Thus, while it is not advisable to do so, standard multiple linear regression may be performed by routine SVGLM. Estimates for any of ten standard models can be computed. Exact, left‑censored, right‑censored, or interval‑censored observations are allowed. (Note that left censoring is the same as interval censoring with left endpoint equal to the left endpoint of the support of the distribution.)
Let η = xTβ be the linear parameterization, where x is a design vector obtained in SVGLM via routine GRGLM (see Chapter 2, “Regression”) from a row of X, and β is a vector of parameters associated with the linear model. Let T denote the random response variable and S(t) denote the probability that T > t. All models considered also allow a fixed parameter wi for observation i (input in column IFIX of X). Use of this parameter is discussed below. There may also be nuisance parameters θ > 0, or σ > 0 to be estimated (along with β) in the various models. Let Φ denote the cumulative normal distribution. The survival models available in SVGLM are:
Model | Name | S(t) |
|---|---|---|
0 | Exponential | |
1 | Linear hazard | |
2 | Log‑normal | |
3 | Normal | |
4 | Log‑logistic | |
5 | Logistic | |
6 | Log least extreme value | |
7 | Least extreme value | |
8 | Log extreme value | |
9 | Extreme value | |
10 | Weibull |
Note that the log‑least‑extreme‑value model is a reparameterization of the Weibull model. Moreover, models 0, 1, 2, 4, 6, 8, and 10 require that T > 0, while all of the remaining models allow any value for T,‑∞ < T < ∞.
Each row in the data matrix can represent a single observation, or, through the use of column IFRQ, it can represent several observations. Classification variables and their products are easily incorporated into the models via the usual GLM type specifications through the use of variables NCLVAR and INDCL, and the model variables NEF, NVEF, and INDEF.
The constant parameter wi is input in X and may be used for a number of purposes. For example, if the parameter in an exponential model is known to depend upon the size of the area tested, volume of a radioactive mass, or population density, etc., then a multiplicative factor of the exponential parameter λ = exp(xβ) may be known apriori. This factor can be input in wi (wi is the log of the factor). An alternate use of wi is as follows: It may be that λ = exp(x1β1 + x2β2), where β2 is known. Letting wi = x2β2, estimates for β1 can be obtained via SVGLM with the known fixed values for β2. Standard methods can then be used to test hypotheses about β2 via computed log‑likelihoods.
Computational details
The computations proceed as follows:
1. The input arguments are checked for consistency and validity.
2. Estimates for the means of the explanatory variables x (as generated from the model specification via GRGLM (see Chapter 2, “Regression”) are computed. Let ƒi denote the frequency of the observation. Means are computed as
3. If INIT = 0, initial estimates of the parameters for all but the exponential models (MODEL = 0, 1) are are obtained as follows:
A. Routine KAPMR is used to compute a nonparametric estimate of the survival probability at the upper limit of each failure interval. (Because upper limits are used, intervaland left‑censored data are taken to be exact failures at the upper endpoint of the failure interval.) The Kaplan‑Meier estimate is computed under the assumption that all failure distributions are identical (i.e., all β’s but the intercept, if present, are assumed to be zero).
B. If INTCEP = 0, a simple linear regression is performed predicting
where t* is computed at the upper endpoint of each failure interval, t* = t in models 3, 5, 7, and 9, and t* = ln(t) in models 2, 4, 6, 8, and 10, and wi is the fixed constant, if present. If INTCEP is zero, α is fixed at zero, and the model
is fit instead of the model above. In this model, the coefficients β are used in place of the location estimate α above. Here, is estimated from the simple linear regression with α = 0.
C. If the intercept is in the model, then in log‑location‑scale models (models 1–8),
and the initial estimate of the intercept, if present, is taken to be .
In the Weibull model,
and the intercept, if present, is taken to be .
Initial estimates of all parameters β, other than the intercept, are taken to be zero.
If no intercept is in the model, the scale parameter is estimated as above, and the estimates from Step B are used as initial estimates for the β’s.
For exponential models (MODEL = 0, 1), the average total time on test statistic is used to obtain an estimate for the intercept. Specifically, let Tt denote the total number of failures divided by the total time on test. The initial estimate for the intercept is then ln(Tt). Initial estimates for the remaining parameters β are taken as zero, and, if MODEL = 1, the initial estimate for the linear hazard parameter θ is taken to be a small positive number. When the intercept is not in the model, the initial estimate for the parameter θ is taken as a small positive number, and initial estimates of the parameters β are computed via multiple linear regression as above.
4. A quasi‑Newton algorithm is used in the initial iterations based upon a Hessian estimate
where
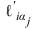
is the partial derivative of the i‑th term in the log‑likelihood with respect to the parameter αj, and αj denotes one of the parameters to be estimated.
When the relative change in the log‑likelihood from one iteration to the next is 0.1 or less, exact second partial derivatives are used for the Hessian so that Newton‑Raphson iteration is used.
If the initial step size results in an increase in the log‑likelihood, the full step is used. If the log‑likelihood decreases for the initial step size, the step size is halved, and a check for an increase in the log‑likelihood performed. Step‑halving is performed (as a simple line search) until an increase in the log‑likelihood is detected, or until the step size is less that 0.0001 (where the initial step size is 1).
5. Convergence is assumed when the maximum relative change in any coefficient update from one iteration to the next is less than EPS, or when the relative change in the loglikelihood from one iteration to the next is less than EPS/100. Convergence is also assumed after MAXIT iterations, or when step halving leads to a step size of less than .0001, with no increase in the log‑likelihood.
6. If requested (INFIN = 0), then the methods of Clarkson and Jennrich (1988) are used to check for the existence of infinite estimates in
As an example of a situation in which infinite estimates can occur, suppose that observation j is right censored with tj > 15 in a normal distribution model in which we fit the mean as
where xj is the observation design vector. If design vector xj for parameter βm is such that xjm = 1 and xim = 0 for all i ≠ j, then the optimal estimate of βm occurs at
leading to an infinite estimate of both βm and ηj. In SVGLM, such estimates may be “computed.”
In all models fit by SVGLM, infinite estimates can only occur when the optimal estimated probability associated with the left- or right‑censored observation is 1. If INFIN = 0, left‑ or right‑censored observations that have estimated probability greater than 0.995 at some point during the iterations are excluded from the log‑likelihood, and the iterations proceed with a log‑likelihood based upon the remaining observations. This allows convergence of the algorithm when the maximum relative change in the estimated coefficients is small and also allows for a more precise determination of observations with infinite
At convergence, linear programming is used to ensure that the eliminated observations have infinite ηi. If some (or all) of the removed observations should not have been removed (because their estimated ηi’s must be finite), then the iterations are restarted with a log‑likelihood based upon the finite ηi observations. See Clarkson and Jennrich (1988) for more details.
When INFIN = 1, no observations are eliminated during the iterations. In this case, when infinite estimates occur, some (or all) of the coefficient estimates will become large, and it is likely that the Hessian will become (numerically) singular prior to convergence.
7. The case statistics are computed as follows:
Let
denote the log‑likelihood of the i‑th observation evaluated at θi, denote the vector of derivatives of with respect to all parameters,
denote the derivative of with respect to η = xTβ, H denote the Hessian, and E denote expectation.
Then, the columns of CASE are:
A. Predicted values are computed as E(T∣x) according to standard formulas. If MODEL is 4 or 8, and if σ ≥ 1, then the expected values cannot be computed because they are infinite.
B. Following Cook and Weisberg (1982), we take the influence (or leverage) of the i‑th observation to be
This quantity is a one‑step approximation to the change in the estimates when the i‑th observation is deleted (ignoring the nuisance parameters).
C. The “residual” is computed as
D. The cumulative hazard is computed at the observation covariate values and, for interval observations, the upper endpoint of the failure interval. The cumulative hazard can also be used as a “residual” estimate. If the model is correct, the cumulative hazards should follow a standard exponential distribution. See Cox and Oakes (1984).
E. The density (for exact failures) or the interval probability (for censored observations) is computed for given x.
Programming Notes
Classification variables are specified by parameters NCLVAR and INDCL. Indicator variables are created for the classification variables using routine GRGLM (see Chapter 2, “Regression”) with IDUMMY = 2.
Examples
Example 1
This example is from Lawless (1982, page 287) and involves the mortality of patients suffering from lung cancer. (The first ten rows of the input data are printed in the output.) An exponential distribution is fit for model
η = μ + β1x3 + β2x4 + β3x5 + αi + γk
where αi is associated with a classification variable with 4 levels, and γk is associated with a classification variable with 2 levels. Note that because the computations are performed in single precision, there will be some small variation in the estimated coefficients across different machine environments.
USE SVGLM_INT
USE WRRRL_INT
IMPLICIT NONE
INTEGER ICEN, ILT, IPAR, IPRINT, IRT, LDCASE, LDCOEF, &
LDCOV, LDX, MAXCL, MODEL, NCLVAR, NCOL, NEF, NOBS
PARAMETER (ICEN=2, ILT=0, IPAR=0, IPRINT=2, IRT=1, LDCASE=40, &
LDCOEF=8, LDCOV=8, LDX=40, MAXCL=6, MODEL=0, &
NCLVAR=2, NCOL=7, NEF=5, NOBS=40)
CHARACTER *6 NUMBER(1)
!
INTEGER IADDS(NOBS), INDCL(NCLVAR), INDEF(5), NCLVAL(NCLVAR), &
NCOEF, NRMISS, NVEF(NEF)
REAL ALGL, CASE(LDCASE,5), CLVAL(MAXCL), COEF(LDCOEF,4), &
COV(LDCOV,LDCOV), GR(LDCOV), X(LDX,NCOL), XMEAN(LDCOV)
!
DATA X/411, 126, 118, 92, 8, 25, 11, 54, 153, 16, 56, 21, 287, &
10, 8, 12, 177, 12, 200, 250, 100, 999, 231, 991, 1, 201, &
44, 15, 103, 2, 20, 51, 18, 90, 84, 164, 19, 43, 340, 231, &
5*0, 1, 16*0, 1, 5*0, 1, 11*0, 7, 6, 7, 4, 4, 7, 7, 8, 6, &
3, 8, 4, 6, 4, 2, 5, 5, 4, 8, 7, 6, 9, 5, 7, 2, 8, 6, 5, 7, &
4, 3, 3, 4, 6, 8, 7, 3, 6, 8, 7, 64, 63, 65, 69, 63, 48, &
48, 63, 63, 53, 43, 55, 66, 67, 61, 63, 66, 68, 41, 53, 37, &
54, 52, 50, 65, 52, 70, 40, 36, 44, 54, 59, 69, 50, 62, 68, &
39, 49, 64, 67, 5, 9, 11, 10, 58, 9, 11, 4, 14, 4, 12, 2, &
25, 23, 19, 4, 16, 12, 12, 8, 13, 12, 8, 7, 21, 28, 13, 13, &
22, 36, 9, 87, 5, 22, 4, 15, 4, 11, 10, 18, 7*1, 7*2, 2*3, &
5*4, 7*1, 4*2, 3*3, 5*4, 21*0, 19*1/
DATA NVEF/1, 1, 1, 1, 1/, INDEF/3, 4, 5, 6, 7/, INDCL/6, 7/
NUMBER(1) = 'NUMBER'
!
CALL WRRRL ('First 10 rows of the input data.', X, &
NUMBER, NUMBER, 10, NCOL, LDX)
!
CALL SVGLM (X, MODEL, ILT, IRT, MAXCL, NCOEF, COEF, ALGL, COV, &
XMEAN, CASE, GR, IADDS, ICEN=ICEN, NCLVAR=NCLVAR, &
INDCL=INDCL, NEF=NEF, NVEF=NVEF, INDEF=INDEF, &
IPRINT=IPRINT, NCLVAL=NCLVAL, CLVAL=CLVAL, &
NRMISS=NRMISS)
!
END
Output
First 10 rows of the input data.
1 2 3 4 5 6 7
1 411.0 0.0 7.0 64.0 5.0 1.0 0.0
2 126.0 0.0 6.0 63.0 9.0 1.0 0.0
3 118.0 0.0 7.0 65.0 11.0 1.0 0.0
4 92.0 0.0 4.0 69.0 10.0 1.0 0.0
5 8.0 0.0 4.0 63.0 58.0 1.0 0.0
6 25.0 1.0 7.0 48.0 9.0 1.0 0.0
7 11.0 0.0 7.0 48.0 11.0 1.0 0.0
8 54.0 0.0 8.0 63.0 4.0 2.0 0.0
9 153.0 0.0 6.0 63.0 14.0 2.0 0.0
10 16.0 0.0 3.0 53.0 4.0 2.0 0.0
Initial Estimates
1 2 3 4 5 6 7 8
-5.054 0.000 0.000 0.000 0.000 0.000 0.000 0.000
Method Iteration Step size Maximum scaled Log
coef. update likelihood
Q-N 0 -224.0
Q-N 1 1.0000 0.9839 -213.4
N-R 2 1.0000 3.603 -207.3
N-R 3 1.0000 10.12 -204.3
N-R 4 1.0000 0.1430 -204.1
N-R 5 1.0000 0.1174E-01 -204.1
Log-likelihood -204.1392
Coefficient Statistics
Standard Asymptotic Asymptotic
Coefficient error z-statistic p-value
1 -1.103 1.314 -0.8939 0.4016
2 -0.540 0.108 -4.995 0.0000
3 -0.009 0.020 -0.459 0.6460
4 -0.003 0.012 -0.291 0.7710
5 -0.363 0.445 -0.816 0.4149
6 0.127 0.486 0.261 0.7939
7 0.869 0.586 1.483 0.1385
8 0.270 0.388 0.695 0.4873
Asymptotic Coefficient Covariance
1 2 3 4 5
1 1.727 -8.1873E-02 -1.9753E-02 -2.2481E-03 -6.5707E-02
2 1.1690E-02 6.4506E-05 2.8955E-04 -3.8734E-04
3 3.8676E-04 -3.9067E-05 -1.2359E-03
4 1.3630E-04 7.5656E-04
5 0.1976
6 7 8
1 -0.1038 -0.1554 -4.2370E-05
2 8.5772E-03 1.8120E-02 6.5272E-03
3 -3.2789E-04 -1.6986E-03 -2.7794E-03
4 -1.6742E-03 6.2668E-04 1.5432E-03
5 9.0035E-02 0.1122 4.3157E-02
6 0.2365 0.1142 -1.3527E-02
7 0.3436 5.1948E-02
8 0.1507
Case Analysis
Cumulative Density or
Predicted Influence Residual Hazard Probability
1 262.7 0.0450 -0.565 1.565 0.0008
2 153.8 0.0042 0.181 0.819 0.0029
3 270.5 0.0482 0.564 0.436 0.0024
4 55.3 0.0844 -0.663 1.663 0.0034
5 61.7 0.3765 0.870 0.130 0.0142
6 230.4 0.0025 -0.108 0.108 0.8972
7 232.0 0.1960 0.953 0.047 0.0041
8 272.8 0.1677 0.802 0.198 0.0030
9 95.9 0.0505 -0.596 1.596 0.0021
10 16.8 0.0005 0.045 0.955 0.0230
11 234.0 0.1911 0.761 0.239 0.0034
12 29.1 0.0156 0.278 0.722 0.0167
13 102.2 0.4609 -1.807 2.807 0.0006
14 34.8 0.0686 0.713 0.287 0.0216
15 5.3 0.0838 -0.521 1.521 0.0415
16 25.7 0.0711 0.533 0.467 0.0244
17 65.6 0.4185 -1.698 2.698 0.0010
18 38.4 0.0886 0.688 0.312 0.0191
19 261.0 0.0155 0.234 0.766 0.0018
20 167.2 0.0338 -0.495 1.495 0.0013
21 85.8 0.0082 -0.166 1.166 0.0036
22 947.8 0.0005 -0.054 1.054 0.0004
23 105.9 0.6402 -2.181 2.181 0.1129
24 305.2 0.5757 -2.247 3.247 0.0001
25 24.6 0.3203 0.959 0.041 0.0390
26 572.8 0.0762 0.649 0.351 0.0012
27 217.5 0.1246 0.798 0.202 0.0038
28 96.6 0.1494 0.845 0.155 0.0089
29 173.4 0.1096 -0.594 0.594 0.5522
30 38.7 0.1928 0.948 0.052 0.0245
31 22.5 0.0040 0.112 0.888 0.0183
32 30.7 0.2270 -0.661 1.661 0.0062
33 20.8 0.0058 0.134 0.866 0.0202
34 54.6 0.1094 -0.648 1.648 0.0035
35 168.6 0.0923 0.502 0.498 0.0036
36 256.8 0.0341 0.361 0.639 0.0021
37 21.9 0.0069 0.134 0.866 0.0192
38 124.3 0.0680 0.654 0.346 0.0057
39 417.9 0.0087 0.186 0.814 0.0011
40 257.1 0.0025 0.101 0.899 0.0016
Last Coefficient Update
1 2 3 4 5 6
-1.031E-05 -1.437E-06 3.098E-07 4.722E-08 -1.844E-05 -1.671E-06
7 8
-2.520E-06 8.139E-06
Covariate Means
1 2 3 4 5 6 7
5.65 56.58 15.65 0.35 0.28 0.12 0.53
Distinct Values For Each Class Variable
Variable 1: 1.0 2.0 3.0 4.0
Variable 2: 0. 1.0
Observation Codes
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Number of Missing Values 0
Example 2
As a second example, the MAXIT = 0 option is used for the model in Example 1 with the coefficients restricted such that μ = ‑1.25, β1 = ‑6, and the remaining 6 coefficients are zero. A chi‑squared statistic with 8 degrees of freedom for testing that the coefficients are specified as above (versus the alternative that they are not as specified) may be computed from the output as
where is output in COV, and g is output in GR. The resulting test statistic (6.107), based upon no iterations, is comparable to the likelihood ratio test statistic that may be computed from the log‑likelihood output in Example 2 (‑206.6835) and the log‑likelihood output in Example 1 (‑204.1392).
Neither test statistic is significant at the α = 0.05 level.
USE IMSL_LIBRARIES
IMPLICIT NONE
INTEGER ICEN, ILT, INIT, IPAR, IPRINT, IRT, LDCASE, &
LDCOEF, LDCOV, LDX, MAXCL, MAXIT, MODEL, NCLVAR, &
NCOL, NEF, NOBS
PARAMETER (ICEN=2, ILT=0, INIT=1, IPAR=0, IPRINT=2,IRT=1, &
LDCASE=40, LDCOEF=8, LDCOV=8, LDX=40, MAXCL=6, &
MAXIT=0, MODEL=0, NCLVAR=2, NCOL=7, NEF=5, NOBS=40)
!
INTEGER IADDS(NOBS), INDCL(NCLVAR), INDEF(5), IRANK, &
NCLVAL(NCLVAR), NCOEF, NRMISS, NVEF(NEF)
REAL ALGL, CASE(LDCASE,5), CHISQ, CLVAL(MAXCL), &
COEF(LDCOEF,4), COV(LDCOV,LDCOV), GR(LDCOV), &
GRD(LDCOV), X(LDX,NCOL), XMEAN(LDCOV)
!
DATA X/411, 126, 118, 92, 8, 25, 11, 54, 153, 16, 56, 21, 287, &
10, 8, 12, 177, 12, 200, 250, 100, 999, 231, 991, 1, 201, &
44, 15, 103, 2, 20, 51, 18, 90, 84, 164, 19, 43, 340, 231, &
5*0, 1, 16*0, 1, 5*0, 1, 11*0, 7, 6, 7, 4, 4, 7, 7, 8, 6, &
3, 8, 4, 6, 4, 2, 5, 5, 4, 8, 7, 6, 9, 5, 7, 2, 8, 6, 5, 7, &
4, 3, 3, 4, 6, 8, 7, 3, 6, 8, 7, 64, 63, 65, 69, 63, 48, &
48, 63, 63, 53, 43, 55, 66, 67, 61, 63, 66, 68, 41, 53, 37, &
54, 52, 50, 65, 52, 70, 40, 36, 44, 54, 59, 69, 50, 62, 68, &
39, 49, 64, 67, 5, 9, 11, 10, 58, 9, 11, 4, 14, 4, 12, 2, &
25, 23, 19, 4, 16, 12, 12, 8, 13, 12, 8, 7, 21, 28, 13, 13, &
22, 36, 9, 87, 5, 22, 4, 15, 4, 11, 10, 18, 7*1, 7*2, 2*3, &
5*4, 7*1, 4*2, 3*3, 5*4, 21*0, 19*1/
DATA NVEF/1, 1, 1, 1, 1/, INDEF/3, 4, 5, 6, 7/, INDCL/6, 7/
!
NCOEF = 8
CALL SSET (NCOEF, 0.0, COEF(3:,1), 1)
CALL ISET (NOBS, 0, IADDS, 1)
COEF(1,1) = -1.25
COEF(2,1) = -0.60
CALL SVGLM (X, MODEL, ILT, IRT, MAXCL, NCOEF, COEF, ALGL, COV, &
XMEAN, CASE, GR(1:1), IADDS, ICEN=ICEN, MAXIT=MAXIT, &
NCLVAR=NCLVAR, INDCL=INDCL, NEF=NEF, NVEF=NVEF,&
INDEF=INDEF, INIT=INIT, IPRINT=IPRINT, &
NCLVAL=NCLVAL, CLVAL=CLVAL)
! Compute Chi-squared
CALL CHFAC (COV, IRANK, COV)
CALL GIRTS (COV, GR, IRANK, GRD, IPATH=2)
!
CHISQ = SDOT(NCOEF,GRD(1:1), 1, GRD(1:1), 1)
WRITE (6,99999) ' Chi-squared statistic with 8 degrees of '// &
'freedom ', CHISQ
!
99999 FORMAT (/, A, G12.4)
END
Output
Log-likelihood -206.6835
Coefficient Statistics
Standard Asymptotic Asymptotic
Coefficient error z-statistic p-value
1 -1.25 1.383 -0.904 0.366
2 -0.60 0.112 -5.365 0.000
3 0.00 0.021 0.000 1.000
4 0.00 0.011 0.000 1.000
5 0.00 0.429 0.000 1.000
6 0.00 0.530 0.000 1.000
7 0.00 0.775 0.000 1.000
8 0.00 0.405 0.000 1.000
Hessian
1 2 3 4 5
1 1.914 -8.1835E-02 -2.3464E-02 -1.1634E-03 -9.0646E-02
2 1.2507E-02 2.0883E-06 3.1320E-04 -5.3147E-04
3 4.6174E-04 -5.5344E-05 -8.1929E-04
4 1.1797E-04 6.0699E-04
5 0.1839
6 7 8
1 -0.1641 -0.1681 7.7768E-02
2 1.0372E-02 1.9269E-02 5.9762E-03
3 5.1246E-04 -1.6419E-03 -4.0106E-03
4 -2.0693E-03 6.9029E-04 1.7020E-03
5 9.9640E-02 0.1191 3.5786E-02
6 0.2808 0.1264 -2.2602E-02
7 0.6003 4.6015E-02
8 0.1641
Estimated Probability (censored) or Estimated Density (non-censored)
1 2 3 4 5 6 7 8
0.0007 0.0029 0.0026 0.0024 0.0211 0.8982 0.0041 0.0021
9 10 11 12 13 14 15 16
0.0024 0.0222 0.0021 0.0151 0.0008 0.0200 0.0433 0.0120
17 18 19 20 21 22 23 24
0.0011 0.0190 0.0015 0.0015 0.0036 0.0004 0.0371 0.0001
25 26 27 28 29 30 31 32
0.0792 0.0015 0.0055 0.0115 0.6424 0.0247 0.0184 0.0042
33 34 35 36 37 38 39 40
0.0163 0.0039 0.0019 0.0021 0.0193 0.0056 0.0011 0.0016
Newton-Raphson Step
1 2 3 4 5 6 7 8
0.171 0.062 -0.011 -0.003 -0.336 0.133 1.297 0.298
Covariate Means
1 2 3 4 5 6 7
5.65 56.58 15.65 0.35 0.28 0.12 0.53
Distinct Values For Each Class Variable
Variable 1: 1.0 2.0 3.0 4.0
Variable 2: 0. 1.0
Observation Codes
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Number of Missing Values 0
Chi-squared statistic with 8 degrees of freedom 6.107
Published date: 03/19/2020
Last modified date: 03/19/2020