Analyzes a balanced complete experimental design for a fixed, random, or mixed model.
Synopsis
#include <imsls.h>
float *imsls_f_anova_balanced (int n_factors, int n_levels[], float y[], int n_random, int index_random_factor[], int n_model_effects, int n_factors_per_effect[], int index_factor_per_effect[], ..., 0)
The type double function is imsls_d_anova_balanced.
Required Arguments
int n_factors
(Input)
Number of factors (number of subscripts) in the model, including
error.
int n_levels[]
(Input)
Array of length n_factors containing
the number of levels for each of the factors.
float y[]
(Input)
Array of length n_levels[0] * n_levels[1]
*.
. .* n_levels[n_factors-1]
containing the responses. y[] must not contain
NaN (not a number) for any of its elements, i.e., missing values are not
allowed.
int n_random
(Input)
For positive n_random, |n_random|
is the number of random factors. For negative n_random,
|n_random| is the number of random effects (sources of
variation).
int index_random_factor[]
(Input)
Index array of length |n_random| containing
either the factor numbers to be considered random (for n_random positive) or
containing the effect numbers to be considered random (for n_random
negative). If n_random = 0, index_random_factor is
not referenced.
int n_model_effects
(Input)
Number of effects (sources of variation) due to the model
excluding the overall mean and error.
int n_factors_per_effect[]
(Input)
Array of length n_model_effects
containing the number of factors associated with each effect in the model.
int
index_factor_per_effect[]
(Input)
Index vector of length n_factors_per_efffect[0] +
n_factors_per_effect[1] + .
. . +
n_factors_per_effect[n_model_effects-1].
The first n_factors_per_effect[0]
elements give the factor numbers in the first effect. The next n_factors_per_effect[1]
elements give the factor numbers in the second effect. The last n_factors_per_effect
[n_model_effects-1] elements give the factor numbers in the last effect.
Main effects must appear before their interactions. In general, an effect
E cannot appear after an effect
F if all of the indices for
E appear also in F.
Return Value
The p-value for the F-statistic.
Synopsis with Optional Arguments
#include <imsls.h>
float *imsls_f_anova_balanced (int n_factors, int n_levels[], float y[], int n_random, int index_random_factor[], int n_model_effects, int n_factors_per_effect[], int index_factor_per_effect[],
IMSLS_ANOVA_TABLE, float **anova_table,
IMSLS_ANOVA_TABLE_USER,
float
anova_table[]
IMSLS_MODEL, int
model,
IMSLS_CONFIDENCE,
float
confidence, IMSLS_VARIANCE_COMPONENTS, float
**variance_components,
IMSLS_VARIANCE_COMPONENTS_USER, float
variance_components[],
IMSLS_EMS, float
**ems,
IMSLS_EMS_USER, float ems[],
IMSLS_Y_MEANS,
float
**y_means,
IMSLS_Y_MEANS_USER,
float
y_means[],
0)
Optional Arguments
IMSLS_ANOVA_TABLE,
float **anova_table,
(Output)
Address of a pointer to an internally allocated array of size 15
containing the analysis of variance table. The analysis of variance statistics
are as follows:
Element Analysis of Variance Statistics
0 Degrees of freedom for the model
1 Degrees of freedom for error
2 Total (corrected) degrees of freedom
3 Sum of squares for the model
4 Sum of squares for error
5 Total (corrected) sum of squares
6 Model mean square
7 Error mean square
8 Overall F-statistic
9 p-value
10 R2 (in percent)
11 adjusted R2 (in percent)
12 estimate of the standard deviation
13 overall mean of Y
14 coefficient of variation (in percent)
IMSLS_ANOVA_TABLE_USER,
float anova_table[]
(Output)
Storage for array anova_table is provided by the user.
See IMSLS_ANOVA_TABLE.
IMSLS_MODEL,
int model, (Input)
Model Option
MODEL Meaning
0 Searle model
1 Scheffe model
For the Scheffe model, effects corresponding to interactions of fixed and random factors have their sum over the subscripts corresponding to fixed factors equal to zero. Also, the variance of a random interaction effect involving some fixed factors has a multiplier for the associated variance component that involves the number of levels in the fixed factors. The Searle model has no summation restrictions on the random interaction effects and has a multiplier of one for each variance component. The default is model = 0.
IMSLS_CONFIDENCE,
float confidence
(Input)
Confidence level for two-sided interval estimates on the variance
components, in percent. confidence
percent confidence intervals are computed, hence, confidence must be in the
interval [0.0, 100.0). confidence often will be 90.0, 95.0, or 99.0.
For
one-sided intervals with confidence level α, α
in the interval [50.0,
100.0),
set confidence = 100.0 - 2.0 * 100.0 - α).
Default:
confidence = 95.0
IMSLS_VARIANCE_COMPONENTS, float **variance_components, (Output) Address of a pointer to an array, variance_components. variance_components is an (n_model_effects + 1) by 9 array containing statistics relating to the particular variance components or effects in the model and the error. Rows of variance_components correspond to the n_model_effects effects plus error.
Element Description
1 Degrees of freedom
2 Sum of squares
3 Mean squares
4 F -statistic
5 p-value for F test
6 Variance component estimate
7 Percent of variance of y explained by random effect
8 Lower endpoint for a confidence interval on the variance component
9 Upper endpoint for a confidence interval on the variance component
Elements 6 through 9 contain NaN (not a number) if the effect is fixed, i.e., if there is no variance component to be estimated. If the variance component estimate is negative, columns 8 and 9 contain NaN.
IMSLS_VARIANCE_COMPONENTS_USER,
float variance_components[]
(Output)
Storage for array variance_components is provided by the
user.
See IMSLS_VARIANCE_COMPONENTS.
IMSLS_EMS,
float **ems, (Output)
Address of a pointer to an internally allocated array of length (n_model_effects + 1) *
(n_model_effects + 2)/2 containing expected mean square coefficients.
Suppose the effects are
A, B, and AB. The ordering of
the coefficients in ems is as follows:
|
|
Error |
AB |
B |
A |
|
A |
ems[0] |
ems[1] |
ems[2] |
ems[2 |
|
B |
ems[4] |
ems[5] |
ems[6] |
|
|
AB |
ems[7] |
ems[8] |
|
|
|
Error |
ems[9] |
|
|
|
IMSLS_EMS_USER,
float ems[]
(Output)
Storage for ems is provided by the
user.
See IMSLS_EMS.
IMSLS_Y_MEANS,
float **y_means
(Output)
Address of a pointer to an internally allocated array of length
(n_levels(0) +
1) * (n_levels (1) + 1)
* .
. . *
(n_levels (n-1) + 1)
containing the subgroup means. Suppose the factors are A, B, and C. The ordering
of the means is grand mean, A means, B means, C means, AB means, AC means, BC
means, and ABC means.
IMSLS_Y_MEANS_USER,
float y_means
(Output)
Storage for y_means is provided by
the user.
See IMSLS_Y_MEANS.
Description
Function imsls_f_anova_balanced analyzes a balanced complete experimental design for a fixed, random, or mixed model. The analysis includes an analysis of variance table, and computation of subgroup means and variance component estimates. A choice of two parameterizations of the variance components for the model can be made.
Scheffé (1959, pages 274−289) discusses the parameterization for model = 1. For example, consider the following model equation with fixed factor A and random factor B:
yijk = m + ai + bj + cij + eijk i = 1, 2, ¼, a; j = 1, 2, ¼, b; k = 1, 2, ¼, n
The fixed effects ai’s are subject to the restriction
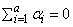
the bj’s are random effects identically and independently distributed
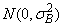
cij are interaction effects each distributed
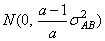
and are subject to the restrictions
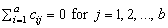
and the eijk’s are errors identically and independently distributed N(0, s2). In general, interactions of fixed and random factors have sums over subscripts corresponding to fixed factors equal to zero. Also in general, the variance of a random interaction effect is the associated variance component times a product of ratios for each fixed factor in the random interaction term. Each ratio depends on the number of levels in the fixed factor. In the earlier example, the random interaction AB has the ratio (a -1)/a as a multiplier of
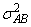
and
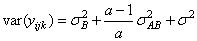
In a three-way crossed classification model, an ABC interaction effect with A fixed, B random, and C fixed would have variance
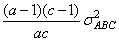
Searle (1971, pages 400−401) discusses the parameterization for model = 0. This parameterization does not have the summation restrictions on the effects corresponding to interactions of fixed and random factors. Also, the variance of each random interaction term is the associated variance component, i.e., without the multiplier. This parameterization is also used with unbalanced data, which is one reason for its popularity with balanced data also. In the earlier example,
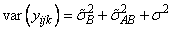
Searle (1971, pages 400−404) compares these two parameterizations. Hocking (1973) considers these different parameterizations and concludes they are equivalent because they yield the same variance-covariance structure for the responses. Differences in covariances for individual terms, differences in expected mean square coefficients and differences in F tests are just a consequence of the definition of the individual terms in the model and are not caused by any fundamental differences in the models. For the earlier two-way model, Hocking states that the relations between the two parameterizations of the variance components are
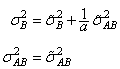
where
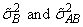
are the variance components in the parameterization with model = 0.
The computations for degrees of freedom and sums of squares are the same regardless of the option specified by model. imsls_f_anova_balanced first computes degrees of freedom and sum of squares for a full factorial design. Degrees of freedom for effects in the factorial design that are missing from the specified model are pooled into the model effect containing the fewest subscripts but still containing the factorial effect. If no such model effect exists, the factorial effect is pooled into error. If more than one such effect exists, a terminal error message is issued indicating a misspecified model.
The analysis of variance method is used for estimating the variance components. This method solves a linear system in which the mean squares are set to the expected mean squares. A problem that Hocking (1985, pages 324−330) discusses is that this method can yield a negative variance component estimate. Hocking suggests a diagnostic procedure for locating the cause of the negative estimate. It may be necessary to re-examine the assumptions of the model.
The percentage of variation explained by each random effect is computed (output in variance_components element 7) as the variance of the associated random effect divided by the variance of y. The two parameterizations can lead to different values because of the different definitions of the individual terms in the model. For example, the percentage associated with the AB interaction term in the earlier two-way mixed model is computed for model = 1 using the formula
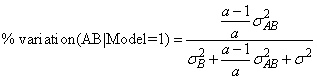
while for the parameterization model = 0, the percentage is computed using the formula
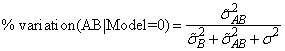
In each case, the variance components are replaced by their estimates (stored in variance_components element 6).
Confidence intervals on the variance components are computed using the method discussed by Graybill (1976, Theorem 15.3.5, page 624, and Note 4, page 620).
Example 1
An analysis of a generalized randomized block design is performed using data discussed by Kirk (1982, Table 6.10-1, pages 293−297). The model is
yijk = m + ai + bj + cij + eijk i = 1, 2, 3, 4; j = 1, 2, 3, 4; k = 1, 2
where yijk is the response for
the k-th experimental unit in block j with treatment
i;
the ai’s are the treatment
effects and are subject to the restriction
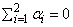
the bj’s are block effects identically and independently distributed
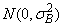
cij are interaction effects each distributed
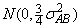
and are subject to the restrictions
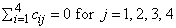
and the eijk’s are errors, identically and independently distributed N(0, s2). The interaction effects are assumed to be distributed independently of the errors.
The data are given in the following table:
|
|
Block | |||
|
Treatment |
1 |
2 |
3 |
4 |
|
1 |
3, 6 |
3, 1 |
2, 2 |
3, 2 |
|
2 |
4, 5 |
4, 2 |
3, 4 |
3, 3 |
|
3 |
7, 8 |
7, 5 |
6, 5 |
6, 6 |
|
4 |
7, 8 |
9, 10 |
10, 9 |
8, 11 |
#include <imsls.h>
#include <stdio.h>
void main()
{
float pvalue = -99.;
int n_levels[] = {4, 4, 2};
int indrf[] = {2, 3};
int nfef[] = {1, 1, 2};
int indef[] = {1, 2, 1, 2};
float y[] = {3.0, 6.0, 3.0, 1.0, 2.0, 2.0, 3.0, 2.0, 4.0, 5.0, 4.0,
2.0, 3.0, 4.0, 3.0, 3.0, 7.0, 8.0, 7.0, 5.0, 6.0, 5.0,
6.0, 6.0, 7.0, 8.0, 9.0, 10.0, 10.0, 9.0, 8.0, 11.0};
float *aov=NULL, *y_means, *variance_components, *ems;
char *aov_labels[] = {
"degrees of freedom for model",
"degrees of freedom for error",
"total (corrected) degrees of freedom",
"sum of squares for model",
"sum of squares for error",
"total (corrected) sum of squares",
"model mean square",
"error mean square",
"F-statistic",
"p-value",
"R-squared (in percent)",
"adjusted R-squared (in percent)",
"est. standard deviation of within error",
"overall mean of y",
"coefficient of variation (in percent)"};
char *ems_labels[] = {
"Effect A and Error",
"Effect A and Effect AB",
"Effect A and Effect B",
"Effect A and Effect A",
"Effect B and Error",
"Effect B and Effect AB",
"Effect B and Effect B",
"Effect AB and Error",
"Effect AB and Effect AB",
"Error and Error"};
char *means_labels[] = {
"Grand mean",
" A means 1",
" A means 2",
" A means 3",
" A means 4",
" B means 1",
" B means 2",
" B means 3",
" B means 4",
"AB means 1 1",
"AB means 1 2",
"AB means 1 3",
"AB means 1 4",
"AB means 2 1",
"AB means 2 2",
"AB means 2 3",
"AB means 2 4",
"AB means 3 1",
"AB means 3 2",
"AB means 3 3",
"AB means 3 4",
"AB means 4 1",
"AB means 4 2",
"AB means 4 3",
"AB means 4 4",};
char *components_labels[] = {
"degrees of freedom for A",
"sum of squares for A",
"mean square of A",
"F-statistic for A",
"p-value for A",
"Estimate of A",
"Percent Variation Explained by A",
"95% Confidence Interval Lower Limit for A",
"95% Confidence Interval Upper Limit for A",
"degrees of freedom for B",
"sum of squares for B",
"mean square of B",
"F-statistic for B",
"p-value for B",
"Estimate of B",
"Percent Variation Explained by B",
"95% Confidence Interval Lower Limit for B",
"95% Confidence Interval Upper Limit for B",
"degrees of freedom for AB",
"sum of squares for AB",
"mean square of AB",
"F-statistic for AB",
"p-value for AB",
"Estimate of AB",
"Percent Variation Explained by AB",
"95% Confidence Interval Lower Limit for AB",
"95% Confidence Interval Upper Limit for AB",
"degrees of freedom for Error",
"sum of squares for Error",
"mean square of Error",
"F-statistic for Error",
"p-value for Error",
"Estimate of Error",
"Percent Explained by Error",
"95% Confidence Interval Lower Limit for Error",
"95% Confidence Interval Upper Limit for Error"};
pvalue = imsls_f_anova_balanced(3, n_levels, y, 2, indrf, 3, nfef, indef,
IMSLS_MODEL, 1,
IMSLS_EMS, &ems,
IMSLS_VARIANCE_COMPONENTS, &variance_components,
IMSLS_Y_MEANS, &y_means,
IMSLS_ANOVA_TABLE, &aov,
0);
printf("p value of F statistic = %f\n", pvalue);
imsls_f_write_matrix("* * * Analysis of Variance * * *", 15, 1, aov,
IMSLS_ROW_LABELS, aov_labels,
IMSLS_WRITE_FORMAT, "%10.5f",
0);
imsls_f_write_matrix("* * * Expected Mean Square Coefficients * * *",
10, 1, ems,
IMSLS_ROW_LABELS, ems_labels,
IMSLS_WRITE_FORMAT, "%6.2f",
0);
imsls_f_write_matrix("* * Analysis of Variance / Variance Components * *",
36, 1,
variance_components,
IMSLS_ROW_LABELS, components_labels,
IMSLS_WRITE_FORMAT, "%10.5f",
0);
imsls_f_write_matrix("means", 25, 1, y_means,
IMSLS_ROW_LABELS, means_labels,
IMSLS_WRITE_FORMAT, "%6.2f",
0);
}
Output
p value of F statistic = 0.000005
* * * Analysis of Variance * * *
degrees of freedom for model 15.00000
degrees of freedom for error 16.00000
total (corrected) degrees of freedom 31.00000
sum of squares for model 216.50000
sum of squares for error 19.00000
total (corrected) sum of squares 235.50000
model mean square 14.43333
error mean square 1.18750
F-statistic 12.15439
p-value 0.00000
R-squared (in percent) 91.93206
adjusted R-squared (in percent) 84.36836
est. standard deviation of within error 1.08972
overall mean of y 5.37500
coefficient of variation (in percent) 20.27395
* * * Expected Mean Square Coefficients * * *
Effect A and Error 1.00
Effect A and Effect AB 2.00
Effect A and Effect B 0.00
Effect A and Effect A 8.00
Effect B and Error 1.00
Effect B and Effect AB 0.00
Effect B and Effect B 8.00
Effect AB and Error 1.00
Effect AB and Effect AB 2.00
Error and Error 1.00
* * Analysis of Variance / Variance Components * *
degrees of freedom for A 3.00000
sum of squares for A 194.50000
mean square of A 64.83334
F-statistic for A 32.87324
p-value for A 0.00004
Estimate of A ..........
Percent Variation Explained by A ..........
95% Confidence Interval Lower Limit for A ..........
95% Confidence Interval Upper Limit for A ..........
degrees of freedom for B 3.00000
sum of squares for B 4.25000
mean square of B 1.41667
F-statistic for B 1.19298
p-value for B 0.34396
Estimate of B 0.02865
Percent Variation Explained by B 1.89655
95% Confidence Interval Lower Limit for B 0.00000
95% Confidence Interval Upper Limit for B 2.31682
degrees of freedom for AB 9.00000
sum of squares for AB 17.75000
mean square of AB 1.97222
F-statistic for AB 1.66082
p-value for AB 0.18016
Estimate of AB 0.39236
Percent Variation Explained by AB 19.48276
95% Confidence Interval Lower Limit for AB 0.00000
95% Confidence Interval Upper Limit for AB 2.75803
degrees of freedom for Error 16.00000
sum of squares for Error 19.00000
mean square of Error 1.18750
F-statistic for Error ..........
p-value for Error ..........
Estimate of Error 1.18750
Percent Explained by Error 78.62069
95% Confidence Interval Lower Limit for Error 0.65868
95% Confidence Interval Upper Limit for Error 2.75057
means
Grand mean 5.38
A means 1 2.75
A means 2 3.50
A means 3 6.25
A means 4 9.00
B means 1 6.00
B means 2 5.13
B means 3 5.13
B means 4 5.25
AB means 1 1 4.50
AB means 1 2 2.00
AB means 1 3 2.00
AB means 1 4 2.50
AB means 2 1 4.50
AB means 2 2 3.00
AB means 2 3 3.50
AB means 2 4 3.00
AB means 3 1 7.50
AB means 3 2 6.00
AB means 3 3 5.50
AB means 3 4 6.00
AB means 4 1 7.50
AB means 4 2 9.50
AB means 4 3 9.50
AB means 4 4 9.50
|
Visual Numerics, Inc. PHONE: 713.784.3131 FAX:713.781.9260 |