Analyzes data from balanced and unbalanced randomized complete-block experiments. Unlike anova_factorial, function rcbd_factorial allows for missing data and one or more locations.
Synopsis
#include <imsls.h>
float *imsls_f_rcbd_factorial (int n_obs, int n_locations, int n_factors, int n_levels[], int model[], float y[],…, 0)
The type double function is imsls_d_rcbd_factorial.
Required Arguments
int n_obs
(Input)
Number of missing and non-missing experimental
observations.
int n_locations
(Input)
Number of locations. n_locations must be
one or greater.
int n_factors
(Input)
Number of factors in the model.
int
n_levels[] (Input)
Array of length n_factors+1. The n_levels[0] through n_levels[n_factors-1] contain the number of levels for
each factor. The last element, n_levels[n_factors],
contains the number of blocks at a location. There must be at least two blocks
and two levels for each factor, i.e., n_levels[i] ≥ 2 for i =0, 1, …, n_factors.
int model[] (Input)
A
n_obs by (n_factors+2) array
identifying the location, block and factor levels associated with each
observation in y. The first
column must contain the location identifier and the second column must contain
the block identifier for the observation associated with that row. The
remaining columns, columns 3 through n_factors+2, should
contain the factor level identifiers in the same order used in n_levels. If
n_locations =1,
the first column is still required, but its contents are ignored.
float y[] (Input)
An
array of length n_obs containing the
experimental observations and any missing values. Missing values are
indicated by placing a NaN (not a number) in y. The NaN value can be set
using either the function imsls_f_machine(6) or imsls_d_machine(6),
depending upon whether single or double precision is being used,
respectively.
Return Value
A pointer to the memory location of a two dimensional, n_anova by 6 array containing the ANOVA table, where:
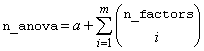 ,
,
 ,
,
and m= n_factors.
Each row in this array contains values for one of the effects in the ANOVA table. The first value in each row, anova_tablei,0 = anova_table[i*6], is the source identifier which identifies the type of effect associated with values in that row. The remaining values in a row contain the ANOVA table values using the following convention:
|
j |
anova_tablei,j = anova_table[i*6+j] |
|
0 |
Source Identifier (values described below) |
|
1 |
Degrees of freedom |
|
2 |
Sum of squares |
|
3 |
Mean squares |
|
4 |
F-statistic |
|
5 |
p-value for this F-statistic |
Note that the p-value for the F-statistic is returned as 0.0 when the value is so small that all significant digits have been lost.
The values for the mean squares, F-statistic and p-value are set to NaN for the residual and corrected total effects.
The Source Identifiers in the first column of anova_tablei,j are the only negative values in anova_table[]. The absolute value of the source identifier is equal to the order of the effect in that row. Main effects, for example, have a source identifier of –1. Two-way interactions use a source identifier of –2, –3 and so on.
|
Source |
|
|
-1 |
Main Effects † |
|
-2 |
Two-Way Interactions ‡ |
|
-3 |
Three-Way Interactions ‡ |
|
. |
. |
|
. |
. |
|
. |
. |
|
-n_factors |
(n_factors)-way Interactions ‡ |
|
-n_factors-1 |
Error Term for Factors and Interactions |
|
-n_factors-2 |
Residual * |
|
-n_factors-3 |
Corrected Total |
Note: The Effects Error Term is equal to the Residual effect if n_locations = 1.
† The number of main effects is equal to n_factors+2 if n_locations > 1, and n_factors +1 if n_locations = 1. The first two rows, anova_table[0] through anova_table[10] are used to represent the location and block effects if n_locations > 1. If n_locations =1, then anova_table[0] through anova_table[5]contain the block effects.
‡ The number of interaction effects for the nth-way interactions is equal to
 .
.
The order of these terms is in ascending order by treatment subscript. The interactions for factor 1 appear first, followed by factor 2, factor 3, and so on.
* The residual term is only produced when there is replication within blocks.
Synopsis with Optional Arguments
#include <imsls.h>
float
*imsls_f_rcbd_factorial (int
n_obs, int
n_locations,
int n_factors,
int n_levels[],
int model[],float y[],
IMSLS_RETURN_USER, float
anova_table[],
IMSLS_N_MISSING, int
*n_missing,
IMSLS_CV, float
*cv,
IMSLS_GRAND_MEAN, float
*grand_mean,
IMSLS_FACTOR_MEANS, float
**factor_means,
IMSLS_FACTOR_MEANS_USER, float
factor_means[],
IMSLS_FACTOR_STD_ERRORS, float
**factor_std_err,
IMSLS_FACTOR_STD_ERRORS_USER, float
factor_std_err[],
IMSLS_TWO_WAY_MEANS, float
**two_way_means,
IMSLS_TWO_WAY_MEANS_USER, float
two_way_means[],
IMSLS_TWO_WAY_STD_ERRORS, float
**two_way_std_err,
IMSLS_TWO_WAY_STD_ERRORS_USER,
float
two_way_std_err[],
IMSLS_TREATMENT_MEANS, float
**treatment_means,
IMSLS_TREATMENT_MEANS_USER, float
treatment_means[],
IMSLS_TREATMENT_STD_ERROR, float
**treatment_std_err,
IMSLS_TREATMENT_STD_ERROR_USER,
float treatment_std_err[]
IMSLS_ANOVA_ROW_LABELS, char
***anova_row_labels,
IMSLS_ANOVA_ROW_LABELS_USER, char
*anova_row_labels[],
0)
Optional Arguments
IMSLS_RETURN_USER,
float
anova_table[] (Output)
User defined n_anova by 6 array for
the anova_table.
IMSLS_N_MISSING,
int *n_missing
(Output)
Number of missing values, if any, found in y. Missing
values are denoted with a NaN (Not a Number) value.
IMSLS_CV, float
*cv
(Output)
Coefficient of Variation computed by:
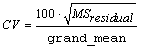 .
.
IMSLS_GRAND_MEAN,
float *grand_mean
(Output)
Mean of all the data across every location.
IMSLS_FACTOR_MEANS,
float
**factor_means (Output)
Address of a pointer to an
internally allocated array of length n_levels[0]+n_levels[1]+…+n_levels[n_factors-1]
containing the factor means.
IMSLS_FACTOR_MEANS_USER,
float
factor_means[] (Output)
Storage for the array factor_means, provided
by the user.
IMSLS_FACTOR_STD_ERRORS,
float
**factor_std_err (Output)
Address of a pointer to an
internally allocated n_factors by 2 array
containing factor standard errors and their associated degrees of freedom.
The first column contains the standard errors for comparing two factor
means and the second its associated degrees of freedom
IMSLS_FACTOR_STD_ERRORS_USER,
float
factor_std_err[] (Output)
Storage for the array factor_std_err,
provided by the user.
IMSLS_TWO_WAY_MEANS,
float
**two_way_means (Output)
Address of a pointer to an
internally allocated one-dimensional array containing the two-way means for all
two by two combinations of the factors. The total length of this array
when n_factors >1
is equal to
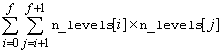 ,
,
where
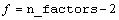
If n_factors = 1, NULL is returned. If n_factors>1, the means would first be produced for all combinations of the first two factors followed by all combinations of the remaining factors using the subscript order suggested by the above formula. For example, if the experiment is a 2x2x2 factorial, the 12 two-way means would appear in the following order: A1B1, A1B2, A2B1, A2B2, A1C1, A1C2, A2C1, A2C2, B1C1, B1C2, B2C1, and B2C2.
IMSLS_TWO_WAY_MEANS_USER,
float two_way_means[]
(Output)
Storage for the array two_way_means,
provided by the user.
IMSLS_TWO_WAY_STD_ERRORS,
float **two_way_std_err
(Output)
Address of a pointer to an internally allocated n_two_way by 2 array
containing factor standard errors and their associated degrees of freedom.,
where
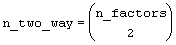
The first column contains the standard errors for comparing two 2-way interaction means and the second its associated degrees of freedom. The ordering of the rows in this array is similar to that used in IMSLS_TWO_WAY_MEANS. For example if n_factors=4, then n_two_way = 6 with the order AB, AC, AD, BC, BD, CD.
IMSLS_TWO_WAY_STD_ERRORS_USER,
float two_way_std_err[]
(Output)
Storage for the array two_way_std_err,
provided by the user.
IMSLS_TREATMENT_MEANS,
float **treatment_means
(Output)
Address of a pointer to an internally allocated array of
size
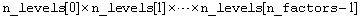
containing the treatment means. The order of the means is organized in ascending order by the value of the factor identifier. For example, if the experiment is a 2x2x2 factorial, the 8 means would appear in the following order: A1B1C1, A1B1C2, A1B2C1, A1B1C2, A2B1C1, A2B1C2, A2B2C1, and A2B2C2.
IMSLS_TREATMENT_MEANS_USER,
float treatment_means[]
(Output)
Storage for the array treatment_means,
provided by the user.
IMSLS_TREATMENT_STD_ERROR,
float **treatment_std_err
(Output)
The array of length 2 containing standard error for comparing
treatments based upon the average number of replicates per treatment and its
associated degrees of freedom.
IMSLS_TREATMENT_STD_ERROR_USER,
float treatment_std_err[]
(Output)
Storage for the array treatment_std_err, provided by the user.
IMSLS_ANOVA_ROW_LABELS,
char ***anova_row_labels
(Output)
Address of a pointer to a pointer to an internally allocated array
containing the labels for each of the n_anova rows of the
returned ANOVA table. The label for the i-th row of the
ANOVA table can be printed with printf("%s", anova_row_labels[i]).
The memory associated with anova_row_labels can be freed with a single call to free(anova_row_labels).
IMSLS_ANOVA_ROW_LABELS_USER,
char *anova_row_labels[]
(Output)
Storage for the array anova_row_labels,
provided by the user. The amount of space required will vary depending
upon the number of factors and n_anova. An
upperbound on the required memory is char *anova_row_labels[100*(n_anova+1)].
Description
The function imsls_f_rcbd_factorial is capable of analyzing randomized complete block factorial experiments replicated in different locations. Missing observations are estimated using the Yates method. Locations, if used, and blocks are treated as random factors. All treatment factors are regarded as fixed effects in the analysis. If n_locations > 1, then blocks are treated as nested within locations and the number of blocks used at each location must be the same.
If n_locations = 1, then the residual mean square is used as the error mean square in calculating the F-tests for all other effects. That is
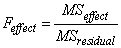 ,
,
when n_locations = 1.
In this case, the residual mean square is calculating by pooling all interactions between treatments and blocks. For example, if treatments are formed from two factors, A and B, then

When n_locations
= 1, then 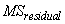 is also used to calculate the standard
errors between means. For example, in a two factor experiment:
is also used to calculate the standard
errors between means. For example, in a two factor experiment:
 ,
,
where
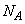 ,
, 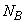 and
and 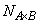
are the number of observations for each level of the effects A, B and their interaction, respectively.
If n_locations > 1, then the error mean square is used as the denominator of the F-test for effects:
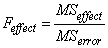 .
.
The error mean square in this calculation is obtained by pooling all interactions between each factor and locations. For example n_locations > 1 and n_factors=2 then:
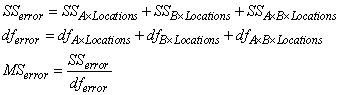
In this case, n_locations > 1, the standard errors for means are calculated using
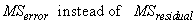
The F-test for differences between locations is calculated using the mean squares for blocks within locations:
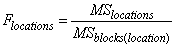
Example
This example is based upon data from an agricultural trial conducted by DOW Agrosciences. This is a three factor, 3x2x2, experiment replicated in two blocks at one location. For illustration, two observations are set to NaN to simulate missing observations.
#include <stdio.h>
#include <math.h>
#include <imsls.h>
int main(){
int n_obs = 24;
int n_locations = 1;
int n_factors = 3;
int n_levels[4] ={3, 2, 2, 2};
int model[]={
1, 1, 1, 1, 1,
1, 2, 1, 1, 1,
1, 1, 1, 1, 2,
1, 2, 1, 1, 2,
1, 1, 1, 2, 1,
1, 2, 1, 2, 1,
1, 1, 1, 2, 2,
1, 2, 1, 2, 2,
1, 1, 2, 1, 1,
1, 2, 2, 1, 1,
1, 1, 2, 1, 2,
1, 2, 2, 1, 2,
1, 1, 2, 2, 1,
1, 2, 2, 2, 1,
1, 1, 2, 2, 2,
1, 2, 2, 2, 2,
1, 1, 3, 1, 1,
1, 2, 3, 1, 1,
1, 1, 3, 1, 2,
1, 2, 3, 1, 2,
1, 1, 3, 2, 1,
1, 2, 3, 2, 1,
1, 1, 3, 2, 2,
1, 2, 3, 2, 2
};
float y[] ={
4.42725419998168950, 2.98526261840015650,
2.12795543670654300, 4.36357164382934570,
2.55254390835762020, 2.78596709668636320,
1.21479606628417970, 2.68143519759178160,
2.47588264942169190, 4.69543695449829100,
5.01306104660034180, 3.01919978857040410,
4.73502767086029050, 0.00000000000000000,
0.00000000000000000, 5.05780076980590820,
5.01421167794615030, 3.61517095565795900,
4.11972457170486450, 4.71947982907295230,
6.51671624183654790, 4.22036057710647580,
4.73365202546119690, 4.68545144796371460
};
int page_width = 132;
int model_order;
int i, n_subscripts, n_anova_table;
char **aov_labels;
char *col_labels[] = {" ", "ID", "df", "SS",
"MS", "F-Test", "P-Value"};
float *anova_table;
/* Compute number of rows in the anova table. */
model_order = n_subscripts = n_factors;
n_anova_table = 3;
for (i=1; i <= model_order; i++){
n_anova_table += imsls_d_binomial_coefficient(n_subscripts, i);
}
/* Set missing observations. */
y[13] = imsls_d_machine(6);
y[14] = imsls_d_machine(6);
anova_table = imsls_f_rcbd_factorial(n_obs, n_locations, n_factors,
n_levels, model, y,
IMSLS_ANOVA_ROW_LABELS, &aov_labels,
0) ;
imsls_page(IMSLS_SET_PAGE_WIDTH, &page_width);
/*
* Print ANOVA table.
*/
imsls_f_write_matrix(" *** ANALYSIS OF VARIANCE TABLE ***",
n_anova_table, 6, anova_table,
IMSLS_ROW_LABELS, aov_labels,
IMSLS_COL_LABELS, col_labels,
IMSLS_WRITE_FORMAT, "%3.0f%3.0f%8.2f%7.2f%7.2f%7.3f",
0);
}
Output
*** ANALYSIS OF VARIANCE TABLE ***
ID df SS MS F-Test P-Value
Blocks -1 1 0.01 0.01 ....... .......
[1] -1 2 14.73 7.37 5.15 0.032
[2] -1 1 0.24 0.24 0.17 0.692
[3] -1 1 0.15 0.15 0.10 0.756
[1]x[2] -2 2 5.79 2.89 2.02 0.188
[1]x[3] -2 2 1.02 0.51 0.36 0.709
[2]x[3] -2 1 0.20 0.20 0.14 0.719
[1]x[2]x[3] -3 2 0.13 0.07 0.05 0.956
Error -4 9 12.88 1.43 ....... .......
Total -6 21 35.15 ....... ....... .......
