Produces summary statistics for a regression model given the information from the fit.
Synopsis
#include <imsls.h>
void imsls_f_regression_summary (Imsls_f_regression *regression_info, ..., 0)
The type double function is imsls_d_regression_summary.
Required Argument
Imsls_f_regression
*regression_info (Input)
Pointer to a structure of type
Imsls_f_regression containing information about the regression fit. See
imsls_f_regression.
Synopsis with Optional Arguments
#include <imsls.h>
void
imsls_f_regression_summary (Imsls_f_regression *regression_info,
IMSLS_INDEX_REGRESSION,
int
idep,
IMSLS_COEF_T_TESTS,
float
**coef_t_tests
IMSLS_COEF_T_TESTS_USER,
float
coef_t_tests[],
IMSLS_COEF_COL_DIM,
int
coef_col_dim,
IMSLS_COEF_VIF,
float
**coef_vif,
IMSLS_COEF_VIF_USER,
float
coef_vif[],
IMSLS_COEF_COVARIANCES,
float
**coef_covariances,
IMSLS_COEF_COVARIANCES_USER,
float
coef_covariances[],
IMSLS_COEF_COV_COL_DIM,
int
coef_cov_col_dim,
IMSLS_ANOVA_TABLE,
float
**anova_table,
IMSLS_ANOVA_TABLE_USER,
float
anova_table[],
0)
Optional Arguments
IMSLS_INDEX_REGRESSION, int idep
(Input)s
Given a multivariate regression fit, this option allows the user to
specify for which regression summary statistics will be computed.
Default:
idep
= 0
IMSLS_COEF_T_TESTS, float
**coef_t_tests (Output)
Address of a pointer to the npar × 4 array
containing statistics relating to the regression coefficients, where npar
is equal to the number of parameters in the model.
Each row (for each dependent variable) corresponds to a coefficient in the model, where npar is the number of parameters in the model. Row i + intcep corresponds to the i-th independent variable, where intcep is equal to 1 if an intercept is in the model and 0 otherwise, for i = 0, 1, 2, …, npar – 1.
The statistics in the columns are as follows:
|
Column |
Description |
|
0 |
coefficient estimate |
|
1 |
estimated standard error of the coefficient estimate |
|
2 |
t-statistic for the test that the coefficient is 0 |
|
3 |
p-value for the two-sided t test |
IMSLS_COEF_T_TESTS_USER, float
coef_t_tests[] (Output)
Storage for array coef_t_tests is
provided by the user. See IMSLS_COEF_T_TESTS.
IMSLS_COEF_COL_DIM, int
coef_col_dim (Input)
Column dimension of coef_t_tests.
Default:
coef_col_dim = 4
IMSLS_COEF_VIF, float
**coef_vif (Output)
Address of a pointer to an internally
allocated array of length npar containing the variance inflation
factor, where npar is the number of parameters. The
i + intcep-th column corresponds to the i-th independent
variable, where i = 0, 1, 2, …, npar - 1,
and intcep is equal to 1 if an intercept is in the model and 0
otherwise.
The square of the multiple correlation coefficient for the i-th regressor after all others can be obtained from coef_vif by
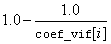
If there is no intercept, or there is an intercept and j = 0, the multiple correlation coefficient is not adjusted for the mean.
IMSLS_COEF_VIF_USER, float
coef_vif[] (Output)
Storage for array coef_t_tests is
provided by the user. See IMSLS_COEF_VIF.
IMSLS_COEF_COVARIANCES, float
**coef_covariances (Output)
An npar by npar
(where npar is equal to the number of parameters in the model) array that
is the estimated variance-covariance matrix of the estimated regression
coefficients when R is nonsingular and is from an unrestricted regression
fit. See “Remarks” for an
explanation of coef_covariances when
R is singular and is from a restricted regression fit.
IMSLS_COEF_COVARIANCES_USER, float
coef_covariances[] (Output)
Storage for coef_covariances
is provided by the user. See IMSLS_COEF_COVARIANCES.
IMSLS_COEF_COV_COL_DIM, int
coef_cov_col_dim (Input)
Column dimension of coef_covariances.
Default:
coef_cov_col_dim
= the number of parameters in the model
IMSLS_ANOVA_TABLE, float
**anova_table (Output)
Address of a pointer to the array
of size 15 containing the analysis of variance table.
|
Row |
Analysis of Variance Statistic |
|
0 |
degrees of freedom for the model |
|
1 |
degrees of freedom for error |
|
2 |
total (corrected) degrees of freedom |
|
3 |
sum of squares for the model |
|
4 |
sum of squares for error |
|
5 |
total (corrected) sum of squares |
|
6 |
model mean square |
|
7 |
error mean square |
|
8 |
overall F-statistic |
|
9 |
p-value |
|
10 |
R2(in percent) |
|
11 |
adjusted R2 (in percent) |
|
12 |
estimate of the standard deviation |
|
13 |
overall mean of y |
|
14 |
coefficient of variation (in percent) |
If the model has an intercept, the regression and total are corrected for the mean; otherwise, the regression and total are not corrected for the mean, and anova_table[13] and anova_table[14] are set to NaN. Note that the p-value is returned as 0.0 when the value is so small that all significant digits have been lost.
IMSLS_ANOVA_TABLE_USER, float
anova_table[] (Output)
Storage for array anova_table is
provided by the user. See IMSLS_ANOVA_TABLE.
Description
Function imsls_f_regression_summary computes summary statistics from a fitted general linear model. The model is y = Xβ + ɛ, where y is the n × 1 vector of responses, X is the n × p matrix of regressors, β is the p × 1 vector of regression coefficients, and ɛ is the n × 1 vector of errors whose elements are each independently distributed with mean 0 and variance σ2. Function regression can be used to compute the fit of the model. Next, imsls_f_regression_summary uses the results of this fit to compute summary statistics, including analysis of variance, sequential sum of squares, t tests, and an estimated variance-covariance matrix of the estimated regression coefficients.
Some generalizations of the general linear model are allowed. If the i-th element of ɛ has variance of
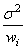
and the weights wi are used in the fit of the model, imsls_f_regression_summary produces summary statistics from the weighted least-squares fit. More generally, if the variance-covariance matrix of ɛ is σ2V, imsls_f_regression_summary can be used to produce summary statistics from the generalized least-squares fit. Function regression can be used to perform a generalized least-squares fit, by regressing y* on X* where y* = (T 1)Ty, X* = (T-1)TX and T satisfies TTT = V.
The sequential sum of squares for the i-th regression parameter is given by
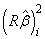
The regression sum of squares is given by the sum of the sequential sums of squares. If an intercept is in the model, the regression sum of squares is adjusted for the mean, i.e.,
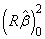
is not included in the sum.
The estimate of σ2 is s2 (stored in anova_table[7]) that is computed as SSE/DFE.
If R is nonsingular, the estimated variance-covariance matrix of
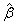
(stored in coef_covariances) is computed by s2R-1(R-1)T.
If R is singular, corresponding to rank(X) < p, a generalized inverse is used. For a matrix G to be a gi (i = 1, 2, 3, or 4) inverse of a matrix A, G must satisfy conditions j (for j ≤ i) for the Moore-Penrose inverse but generally must fail conditions k (for k > i). The four conditions for G to be a Moore-Penrose inverse of A are as follows:
1. AGA = A.
2. GAG = G.
3. AG is symmetric.
4. GA is symmetric.
In the case where R is singular, the method for obtaining coef_covariances follows the discussion of Maindonald (1984, pp. 101–103). Let Z be the diagonal matrix with diagonal elements defined by the following:
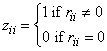
Let G be the solution to RG = Z obtained by setting the i-th ({i : rii = 0}) row of G to 0. Argument coef_covariances is set to s2GGT. (G is a g3 inverse of R, represented by,
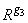
the result
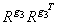
is a symmetric g2 inverse of RTR = XTX. See Sallas and Lionti 1988.)
Note that argument coef_covariances can be used only to get variances and covariances of estimable functions of the regression coefficients, i.e., nonestimable functions (linear combinations of the regression coefficients not in the space spanned by the nonzero rows of R) must not be used. See, for example, Maindonald (1984, pp. 166–168) for a discussion of estimable functions.
The estimated standard errors of the estimated regression coefficients (stored in Column 1 of coef_t_tests) are computed as square roots of the corresponding diagonal entries in coef_covariances.
For the case where an intercept is in the model, put 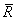 equal to the matrix R with
the first row and column deleted. Generally, the variance inflation factor (VIF)
for the i-th regression
coefficient is computed as the product of the i-th diagonal
element of RTR and the
i-th
diagonal element of its computed inverse. If an intercept is in the model, the
VIF for those coefficients not corresponding to the intercept uses the diagonal
elements of
equal to the matrix R with
the first row and column deleted. Generally, the variance inflation factor (VIF)
for the i-th regression
coefficient is computed as the product of the i-th diagonal
element of RTR and the
i-th
diagonal element of its computed inverse. If an intercept is in the model, the
VIF for those coefficients not corresponding to the intercept uses the diagonal
elements of 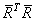 (see Maindonald 1984,
p. 40).
(see Maindonald 1984,
p. 40).
Remarks
When R is nonsingular and comes from an unrestricted regression fit, coef_covariances is the estimated variance-covariance matrix of the estimated regression coefficients, and coef_covariances = (SSE/DFE) (RTR). Otherwise, variances and covariances of estimable functions of the regression coefficients can be obtained using coef_covariances, and coef_covariances = (SSE/DFE) (GDGT). Here, D is the diagonal matrix with diagonal elements equal to 0 if the corresponding rows of R are restrictions and with diagonal elements equal to 1 otherwise. Also, G is a particular generalized inverse of R.
Example
#include <imsls.h>
int main()
{
#define INTERCEPT 1
#define N_INDEPENDENT 4
#define N_OBSERVATIONS 13
#define N_COEFFICIENTS (INTERCEPT + N_INDEPENDENT)
#define N_DEPENDENT 1
Imsls_f_regression *regression_info;
float *anova_table, *coef_t_tests, *coef_vif,
*coefficients, *coef_covariances;
float x[][N_INDEPENDENT] = {
7.0, 26.0, 6.0, 60.0,
1.0, 29.0, 15.0, 52.0,
11.0, 56.0, 8.0, 20.0,
11.0, 31.0, 8.0, 47.0,
7.0, 52.0, 6.0, 33.0,
11.0, 55.0, 9.0, 22.0,
3.0, 71.0, 17.0, 6.0,
1.0, 31.0, 22.0, 44.0,
2.0, 54.0, 18.0, 22.0,
21.0, 47.0, 4.0, 26.0,
1.0, 40.0, 23.0, 34.0,
11.0, 66.0, 9.0, 12.0,
10.0, 68.0, 8.0, 12.0};
float y[] = {78.5, 74.3, 104.3, 87.6, 95.9, 109.2,
102.7, 72.5, 93.1, 115.9, 83.8, 113.3, 109.4};
char *anova_row_labels[] = {
"degrees of freedom for regression",
"degrees of freedom for error",
"total (uncorrected) degrees of freedom",
"sum of squares for regression",
"sum of squares for error",
"total (uncorrected) sum of squares",
"regression mean square",
"error mean square", "F-statistic",
"p-value", "R-squared (in percent)",
"adjusted R-squared (in percent)",
"est. standard deviation of model error",
"overall mean of y",
"coefficient of variation (in percent)"};
/* Fit the regression model */
coefficients = imsls_f_regression(N_OBSERVATIONS, N_INDEPENDENT,
(float *)x, y,
IMSLS_REGRESSION_INFO, ®ression_info,
0);
/* Generate summary statistics */
imsls_f_regression_summary (regression_info,
IMSLS_ANOVA_TABLE, &anova_table,
IMSLS_COEF_T_TESTS, &coef_t_tests,
IMSLS_COEF_VIF, &coef_vif,
IMSLS_COEF_COVARIANCES, &coef_covariances,
0);
/* Print results */
imsls_f_write_matrix("* * * Analysis of Variance * * *\n", 15, 1,
anova_table,
IMSLS_ROW_LABELS, anova_row_labels,
IMSLS_WRITE_FORMAT, "%10.2f", 0);
imsls_f_write_matrix("* * * Inference on Coefficients * * *\n",
N_COEFFICIENTS, 4, coef_t_tests,
IMSLS_WRITE_FORMAT, "%10.2f", 0);
imsls_f_write_matrix("* * * Variance Inflation Factors * * *\n",
N_COEFFICIENTS, 1, coef_vif,
IMSLS_WRITE_FORMAT, "%10.2f", 0);
imsls_f_write_matrix("* * * Variance-Covariance Matrix * * *\n",
N_COEFFICIENTS, N_COEFFICIENTS,
coef_covariances,
IMSLS_WRITE_FORMAT, "%10.2f", 0);
}
Output
* * * Analysis of Variance * * *
degrees of freedom for regression 4.00
degrees of freedom for error 8.00
total (uncorrected) degrees of freedom 12.00
sum of squares for regression 2667.90
sum of squares for error 47.86
total (uncorrected) sum of squares 2715.76
regression mean square 666.97
error mean square 5.98
F-statistic 111.48
p-value 0.00
R-squared (in percent) 98.24
adjusted R-squared (in percent) 97.36
est. standard deviation of model error 2.45
overall mean of y 95.42
coefficient of variation (in percent) 2.56
* * * Inference on Coefficients * * *
1 2 3 4
1 62.41 70.07 0.89 0.40
2 1.55 0.74 2.08 0.07
3 0.51 0.72 0.70 0.50
4 0.10 0.75 0.14 0.90
5 -0.14 0.71 -0.20 0.84
* * * Variance Inflation Factors * * *
1 10668.53
2 38.50
3 254.42
4 46.87
5 282.51
* * * Variance-Covariance Matrix * * *
1 2 3 4 5
1 4909.95 -50.51 -50.60 -51.66 -49.60
2 -50.51 0.55 0.51 0.55 0.51
3 -50.60 0.51 0.52 0.53 0.51
4 -51.66 0.55 0.53 0.57 0.52
5 -49.60 0.51 0.51 0.52 0.50
