clusterHierarchical¶

Performs a hierarchical cluster analysis given a distance matrix.
Synopsis¶
clusterHierarchical (dist)
Required Arguments¶
- float
dist[[]](Input/Ouput) - An
nptbynptsymmetric matrix containing the distance (or similarity) matrix.distis a symmetric matrix. On input, only the upper triangular part needs to be present. The functionclusterHierarchicalsaves the upper triangular part ofdistin the lower triangle. On return fromclusterHierarchical, the upper triangular part ofdistis restored, and the matrix is made symmetric.
Optional Arguments¶
method, int (Input)Option giving the clustering method to be used.
methodMethod 0 Single linkage (minimum distance) 1 Complete linkage (maximum distance) 2 Average distance within (average distance between objects within the merged cluster) 3 Average distance between (average distance between objects in the two clusters) 4 Ward’s method (minimize the within-cluster sums of squares). For Ward’s method, the elements of distare assumed to be Euclidean distances.Default:
method= 0.transformation, int (Input)Option giving the method to be used for clustering.
methodMethod 0 No transformation is required. The elements of distare distances.1 Convert similarities to distances by multiplication by -1.0. 2 Convert similarities (usually correlations) to distances by taking the reciprocal of the absolute value. Default:
transformation= 0.clusters,clevel,iclson,icrson(Output)- Argument
clevelis an array of lengthnpt- 1 containing the level at which the clusters are joined.clevel[k-1] contains the distance (or similarity) level at which clusternpt+ k was formed. If the original data indistwas transformed via the optional argumenttransformation, the inverse transformation is applied to the values inclevelprior to exit fromclusterHierarchical. Argumenticlsonis an array of lengthnpt‑ 1 containing the left sons of each merged cluster. Argumenticrsonis an array of lengthnpt- 1 containing the right sons of each merged cluster. Clusternpt+ k is formed by merging clustersiclson[k‑1] andicrson[k‑1].
Description¶
Function clusterHierarchical conducts a hierarchical cluster analysis
based upon the distance matrix, or by appropriate use of the
transformation optional argument, based upon a similarity matrix. Only
the upper triangular part of the matrix dist is required as input to
clusterHierarchical.
Hierarchical clustering in clusterHierarchical proceeds as follows.
Initially, each data point is considered to be a cluster, numbered 1 to n
= npt.
- If the data matrix contains similarities, they are converted to distances
by the method specified by
transformation. Set \(k=1\). - A search is made of the distance matrix to find the two closest clusters.
These clusters are merged to form a new cluster, numbered n + k. The
cluster numbers of the two clusters joined at this stage are saved in
icrsonandiclson, and the distance measure between the two clusters is stored inclevel. - Based upon the method of clustering, updating of the distance measure in
the row and column of
distcorresponding to the new cluster is performed. - Set \(k=k+1\). If \(k<n\), go to Step 2.
The five methods differ primarily in how the distance matrix is updated
after two clusters have been joined. The method optional argument
specifies how the distance of the cluster just merged with each of the
remaining clusters will be updated. Function clusterHierarchical allows
five methods for computing the distances. To understand these measures,
suppose in the following discussion that clusters “A” and “B”
have just been joined to form cluster “Z”, and interest is in
computing the distance of Z with another cluster called “C”.
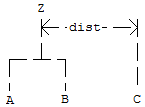
method |
Method |
|---|---|
| 0 | Single linkage method. The distance from Z to C is the minimum of the distances (A to C, B to C). |
| 1 | Complete linkage method. The distance from Z to C is the maximum of the distances (A to C, B to C). |
| 2 | Average-distance-within-clusters method. The distance from Z to C is the average distance of all objects that would be within the cluster formed by merging clusters Z and C. This average may be computed according to formulas given by Anderberg (1973, page 139). |
| 3 | Average-distance-between-clusters method. The distance from Z to C is the average distance of objects within cluster Z to objects within cluster C. This average may be computed according to methods given by Anderberg (1973, page 140). |
| 4 | Ward’s method. Clusters are formed so as to minimize the increase in the within-cluster sums of squares. The distance between two clusters is the increase in these sums of squares if the two clusters were merged. A method for computing this distance from a squared Euclidean distance matrix is given by Anderberg (1973, pages 142-145). |
In general, single linkage will yield long thin clusters while complete linkage will yield clusters that are more spherical. Average linkage and Ward’s linkage tend to yield clusters that are similar to those obtained with complete linkage.
Function clusterHierarchical produces a unique representation of the
binary cluster tree via the following three conventions; the fact that the
tree is unique should aid in interpreting the clusters. First, when two
clusters are joined and each cluster contains two or more data points, the
cluster that was initially formed with the smallest level (in clevel)
becomes the left son. Second, when a cluster containing more than one data
point is joined with a cluster containing a single data point, the cluster
with the single data point becomes the right son. Finally, when two clusters
containing only one object are joined, the cluster with the smallest cluster
number becomes the right son.
Comments¶
- The clusters corresponding to the original data points are numbered from
1 to
npt. Thenpt- 1 clusters formed by merging clusters are numberednpt+ 1 tonpt+ (npt- 1). - Raw correlations, if used as similarities, should be made positive and
transformed to a distance measure. One such transformation can be
performed by specifying optional argument
transformation, withtransformation= 2 inclusterHierarchical. - The user may cluster either variables or observations in
clusterHierarchicalsince a dissimilarity matrix, not the original data, is used. Function dissimilarities may be used to compute the matrixdistfor either the variables or observations.
Example¶
In the following example, the average distance within clusters method is
used to perform a hierarchical cluster analysis of the Fisher Iris data.
Function dataSets (see Chapter
15, Utilities) is first used to obtain the Fisher Iris
data. The example is typical in that after the program obtains the data,
function dissimilarities computes the distance
matrix (dist) prior to calling clusterHierarchical.
from __future__ import print_function
from numpy import *
from pyimsl.stat.clusterHierarchical import clusterHierarchical
from pyimsl.stat.dataSets import dataSets
from pyimsl.stat.dissimilarities import dissimilarities
iscale = 1
ind = (1, 2, 3, 4)
x = dataSets(3)
dist = dissimilarities(x,
index=ind,
scale=iscale)
clusters = {}
clusterHierarchical(dist,
clusters=clusters,
method=2)
clevel = clusters['clevel']
for i in range(0, 149, 15):
print("%6.2f\t" % clevel[i], end=' ')
print()
iclson = clusters['iclson']
for i in range(0, 149, 15):
print("%6i\t" % iclson[i], end=' ')
print()
icrson = clusters['icrson']
for i in range(0, 149, 15):
print("%6i\t" % icrson[i], end=' ')
Output¶
0.00 0.17 0.23 0.27 0.31 0.37 0.41 0.48 0.60 0.78
143 152 100 127 131 198 186 218 261 249
102 29 56 124 108 91 212 243 266 262